INTRODUCTION
The genus Leptosphaerulina was first introduced by McAlpine in 1902, by designating Leptosphaerulina australis as the type species [1]. In 1961, Graham and Luttrell [2] described three existing species and three new species of Leptosphaerulina found on forage plants; the pathogenic Leptosphaerulina arachidicola, L. briosiana, and L. trifolii and the saprobic L. australis, L. argentinensis, and L. americana. However, Irwin and Davis [3] reported that L. argentinensis is pathogenic to Stylosanthes guianensis. Leptosphaerulina species have ostiolate, papillate ascomata, which are immersed to erumpent, and bitunicate asci that are distinctly saccate. The ascospores are oblong to cylindrical, generally muriform, and mostly hyaline, but become brown at maturity [2, 4–6]. The type species of the genus, L. australis, was found on apricot leaves (Prunus armeniaca L.) and has also been reported to be associated with Dolichos Poa, Lolium and Vitis spp. [1, 2]. In the United States, L. australis has been shown to colonize many species from the following genera: Agrostis Festuca, Ligustrum, Lolium, Panicum, Poa, Rosa, and Trifolium [2, 7, 8].
In the present study, a fungal strain, designated KNU16-004, which was isolated from field soil sample, was identified as L. australis based on morphological characterization and phylogenetic analysis using a concatenated dataset of the partial ribosomal large subunit (LSU), internal transcribed spacer (ITS) region, and β-tubulin (Tub2) gene sequences.
MATERIALS and METHODS
Sampling and fungi isolation
Fungal strain KNU16-004 was isolated from a field soil sample collected in Seoul, Korea (N 37°30'59.5", E 127°07'12.6"). The soil sample was collected from the ground, air-dried, and then stored in a plastic bag at 4°C until analysis. The fungus was isolated as previously described [9]. One gram of the soil sample was suspended in 10 mL of sterile distilled water, and the suspension was then vortexed and serially diluted. One hundred microliters of each dilution were spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates, which were incubated at 25°C for 3~7 days until fungal colony growth was observed. Single colonies from these plates were purified by transferring them onto fresh plates and incubating the plates under the same conditions. The pure cultures were routinely cultured on PDA agar at 25°C and maintained as a glycerol suspension (20%, w/v) at −70°C.
For morphological analysis, isolate KNU16-004 was cultured on PDA and incubated at 25°C in the dark for 7 days. Following incubation, colony characteristics such as colour, size, and shape were recorded. In addition, growth was examined on malt extract agar (MEA), synthetic low-nutrient agar (SNA), and oatmeal agar (OA) plates. In order to observe ascomata, ascospores, and asci, the colonies were exposed to illumination with the near-UV light on a 12-hr diurnal cycle. After 7 days of incubation, samples were collected daily for one week and observed for the presence of ascospores using a model BX-50 light microscope (Olympus, Tokyo, Japan).
DNA extraction, PCR, and sequencing analysis
For phylogenic analysis, the genomic DNA of KNU16-004 was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s instructions. Primers ITS1F and ITS4 were used to amplify the ITS regions including the 5.8S rRNA region [10]. Primers LROR [11] and LR5 [12] were used for LSU amplification and primers Btub2Fd and Btub4Rd [13] for amplification of the partial Tub2 gene region. The amplified PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced (SolGent, Daejeon, Korea). Phylogenetic neighbors were identified and similarities with closely related species were calculated using the BLAST search program on the National Center for Biotechnology Information (NCBI) website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The resulting sequences of KNU16-004 have been deposited in the NCBI GenBank/EMBL/DDBJ databases with accession numbers LC420014 for LSU rDNA, LC420015 for ITS rDNA, and LC420016 for Tub2.
Phylogenetic analysis
The LSU, ITS, and Tub2 gene sequences were aligned using the Clustal X computer program [14]. Phylogenetic trees were constructed using a concatenated dataset of partial LSU, ITS, and Tub2 gene sequences, assuming that analysis using a longer sequence would result in better resolution and reliability. Nine related taxa were obtained from GenBank and edited using the BioEdit program [15]; these strains are listed in Table 1. Multiple sequence alignments were performed using the Clustal X program. Gaps and the 5′ and 3′ ends of the alignments were manually edited in BioEdit [15]. Evolutionary distance matrices were generated as described by Kimura [16] and phylogenetic trees were constructed using the neighbor-joining [17], maximum-likelihood [18], and maximum-parsimony [19] algorithms in the MEGA7 program [20], with bootstrap values calculated based on 1,000 replications.
RESULTS and DISCUSSION
Morphology of isolate KNU16-004
At 25°C, isolate KNU16-004 formed flat colonies with sparse aerial mycelium and smooth and lobate margins. After 7 days of incubation, the growth diameter reached 22 mm on PDA, MEA, and OA agar, but only 12 mm on SNA agar (Fig. 1A~1D). Mycelium initially white and then changed to deep black from the center of the colony. The surface of the colony was covered by short, white aerial mycelium, the reverse side of the colony was initially black, and the end of the colony reached the edge of the PDA plate within 21 days of incubation; ascospores and asci were produced on PDA agar after exposure to the above-mentioned illumination and observed under a light microscope. The ascospores were ellipsoid to oblong with a gelatinous sheath, 30.8~34.9 × 8.1~15.8 μm in size, and the asci were bitunicate and thick-walled, 73.5~87.5 × 23.7~37.9 μm in size (Fig. 1F, 1H). As shown in Table 2, these morphological characteristics of isolate KNU16-004 were completely consistent with those previously reported for L. australis [21, 22].
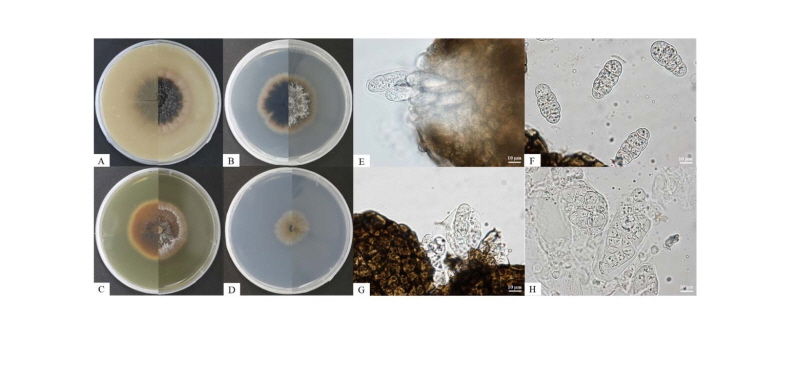
Fig. 1. Culture and morphological characteristics of Leptosphaerulina australis KNU16-004. A, Colony on oatmeal agar; B, Colony on potato dextrose agar; C, Colony on malt extract agar; D, Colony on synthetic low-nutrient agar; E, G, Colony sporulating on ascomata; F, Ascospores; H, Asci (scale bar = 10 μm).
|
Table 2. Morphological characteristics of KNU16-004 isolated in this study and comparison with previously reported isolates 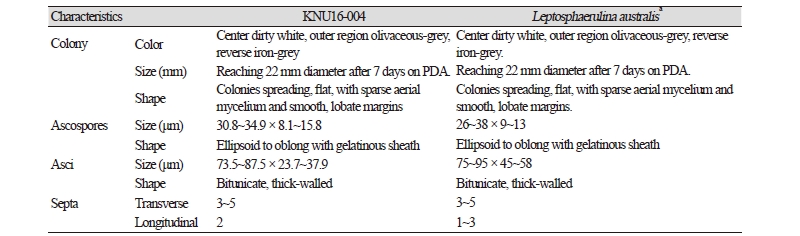
|
|
|
PDA, potato dextrose agar. aSource of description [21, 22]. |
|
Molecular phylogeny of isolate KNU16-004
To identify the isolated fungal strain at the species level, the nucleotide sequences of the LSU, ITS, and Tub2 (890 bp, 519 bp, and 345 bp, respectively) were obtained and compared with the GenBank database using BLAST. The LSU sequence of KNU16-004 exhibited high similarities (99.8~100%) with various L. australis strains, the ITS sequence shared 100% identity with various L. australis strains, and the Tub2 sequence was identical (100%) to L. australis strain CBS 939.69. Phylogenetic analysis based on a concatenated alignment of the LSU, ITS, and Tub2 nucleotide sequences performed using the neighbor-joining, maximum-likelihood maximum-parsimony methods revealed that the strain KNU16-004 belongs to the genus Leptosphaerulina and is grouped with two L. australis strains with a high bootstrap value of 100% (Fig. 2).
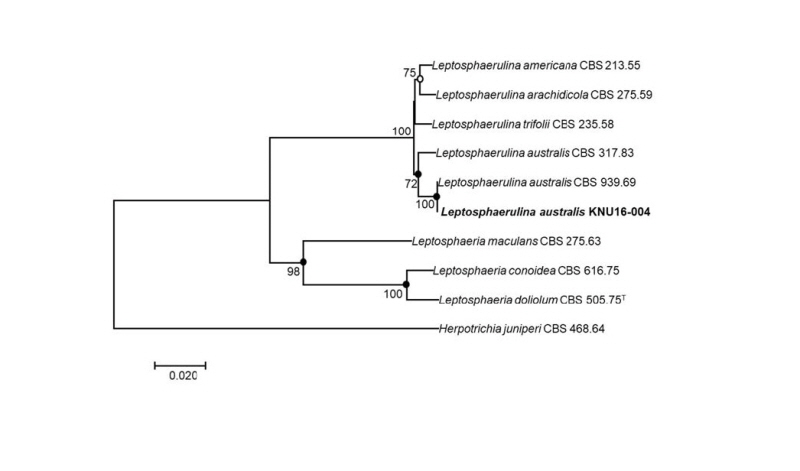
Fig. 2. Neighbor-joining phylogenetic tree based on a concatenated alignment of the large subunit, internal transcribed spacer, and β-tubulin sequences. The phylogenic analysis shows the position of Leptosphaerulina australis KNU16-004 among related Leptosphaerulina spp. strains. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. Filled circles indicate that the corresponding nodes were also recovered in trees generated with the maximum-likelihood and maximum-parsimony algorithms. Open circles indicate that the corresponding nodes were also recovered in the tree generated with the maximum-likelihood algorithm. The tree was rooted using Herpotrichia juniperi CBS 468.64 as an outgroup. Bar, 0.02 substitutions per nucleotide position. CBS, Westerdijk Fungal Biodiversity Institute (formerly CBSKNAW), Utrecht, The Netherlands.
Pathogenicity status of L. australis is uncertain [23]. However, L. australis has been detected in imported agricultural produce, namely on Brassica oleracea var. capitate leaf spots [21], thus, it could be relevant to agricultural product trade. Further studies will be needed to investigate its potential pathogenicity and identify the relative susceptibility of available cultivars. This is the first report of Leptosphaerulina australis in Korea.