INTRODUCTION
The Mortierellales are a species-rich order of early diverging fungi, containing nearly 100 validated species. In the family Mortierellaceae alone, 13 genera have been described, out of which 6 are currently considered valid [1, 2]. The genus Mortierella has been introduced to accommodate the type species Mortierella polycephala initially isolated from a mushroom [3], while a majority of the species is saprobic soil-inhabiting fungi. Many species of Mortierella are widespread in temperate zones, with several ecological factors, and occur in a wide range of habitats [4]. Therefore it is referred to as one of the 10 most frequently detected fungal genera in environmental sequencing projects (). Interestingly, many members of Mortierella exhibit the ability to produce polyunsaturated fatty acids or to convert organic compounds, rendering them attractive for biotechnological studies [5, 6]. In addition, some species potentially play a crucial role in the formation of hydroxylated metabolites, not only rendering them susceptible to degrading for 2,4- dichlorophenol, 2,4-dichlorophenoxyacetic acid, PCNB, and isoproturon, but also remedying the soil contaminated with organochlorine pesticides [7].
In a morphology-based taxonomy [8], Mortierella was further subdivided into 9 sections, but this classical division was mostly in conflict with the DNA sequence-based molecular phylogenies [9, 10]. Instead, Petkovits et al. [9] suggested 12 phylogenetic clades, which were later reclassified into 7 major groups through a more comprehensive study [10]. As a result of recent investigations on fungal biodiversity in Korea over the last 5 years, 5 new species of Mortierella have been recorded from Korean soil samples, namely, M. alpina [11], M. zychae, M. ambigua, M. indohii [12], and M. oligospora [13]. In addition, a new species, Mortierella fluviae [14], along with M. minutissima [15], have been isolated from freshwater environments. However, given the currently accepted species number in this genus and the fact that Mortierella is a ubiquitous group very frequently isolated worldwide, it is quite probable that additional undiscovered species exist in Korea. In the present study, 3 previously unreported Mortierella species were isolated from Korean riverside soils. Herein, we present their morphological characteristics and molecular phylogenetic relationships with the other members of Mortierella.
MATERIALS AND METHODS
Fungal isolates
In 2017 and 2018, 3 species of Mortierella were isolated from Korean riverside. Each sample was collected into a 15 mL conical tube and transferred to laboratory on ice. The samples were then diluted conventionally and inoculated on potato dextrose agar (PDA; Difco, Detroit, MI, USA) with 100 µg/L streptomycin for 2 days at 25°C in dark. When fungal hyphal growth was observed under a microscope, its tip was transferred to a new PDA plate for morphological examination and genomic DNA extraction. Cultivation was performed at 25°C for 3~20 days depending upon the requirements for sporulation. Information of all isolates used in the present study has been shown in Table 1.
|
Table 1. Information of Mortierella strains isolated from riverside soils in Korea 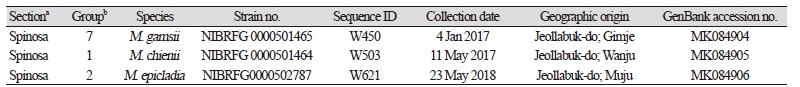
|
|
|
a,bSubdivision of the genus Mortierella, based on amorphology [8] and bphylogeny [10].% |
|
Cultural and morphological characteristics
Cultural characteristics were investigated 3 days after inoculating them at 28°C in dark, on 3 different media, PDA (Difco), V8 agar (V8A; 8% V8 juice [v/v] and 1.5% agar [w/v] adjusted to pH 6.0 using 10 N NaOH), and corn meal agar (CMA; Difco). For microscopy, fungal structures formed on the 3 different media were transferred to a drop of distilled water on a microscope slide and covered with a cover slip. The slides were then examined and photographed using an Olympus BX53F microscope (Olympus, Tokyo, Japan), equipped with a DigiRetina 16M digital camera (Tucsen, Fuzhou, China). At least 50 units for each fungal structure were measured.
Phylogenetic analysis
In total, 5~10 mg of mycelia were ground in a mixer mill (MM2; Retsch, Hann, Germany), using 170 mg of glass beads (Bio Spec Products, Bartlesville, OK, USA) with 1 mm diameter, per sample. Genomic DNA was extracted using the MagListo 5M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) following the manufacturer's instructions. PCR amplification of the complete internal transcribed spacer (ITS) rDNA region was performed with the primer set, ITS1 and ITS4 [16]. Amplicons were visualized on 1.2% agarose gel, purified using an AccuPrep PCR Purification Kit (Bioneer), and sequenced by a DNA sequencing service (Macrogen, Seoul, Korea), with the same primers used for amplification. The ITS rDNA sequences were edited using the DNAStar software package version 5.05 (DNASTAR, Madison, WI, USA). Alignment was performed using MAFFT 7 [17] with the Q-INS-i algorithm [18]. To construct a global phylogeny of Mortierella species, the previously published sequences of the type or authentic strains of Mortierella species, as well as the previously recorded species in Korea, were retrieved from NCBI GenBank. Minimum evolution (ME) tree was constructed with MEGA 6.0 [19], using the Tamura-Nei substitution model and performing 1,000 bootstrap replicates. All the other parameters were set to default. For maximum likelihood (ML) analyses, 1,000 rounds of random addition of sequences as well as 1,000 fast bootstrap replicates were performed using RAxML 7.0.3 [20] as implemented in RAxMLGUI 1.3 [21] using the GTRCAT variant.
RESULTS AND DISCUSSION
The complete ITS rDNA sequences were employed to reconstruct a phylogenetic relationship between Mortierella species. The final alignment displayed 809 total characters, including 540 variable characters, 489 of which were parsimony-informative. The dataset revealed no significant conflicts in the topologies derived from ME and ML analyses. Thus, with unsupported replacements of a few internal branches within each group and somewhat lower bootstrapping values in ML, only the tree from the ME inference has been shown in Fig. 1. The overall topology and significant groupings were coherent with a previous study by Wagner et al. [10]. But in case of our tree, Groups 1 and 6 were paraphyletic, which match the groupings of Petkovits et al. [9]. Monophylies of Groups 2, 3, and 5 were highly supported in both ME and ML analyses, while those of Groups 4 and 7 received no convincing support.
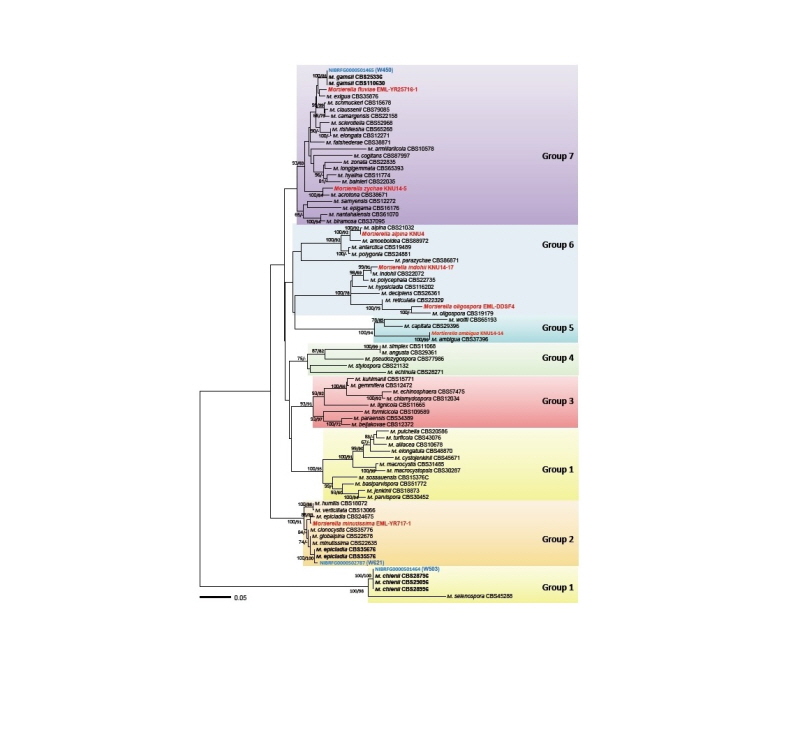
Fig. 1. Minimum evolution tree based on the internal transcribed spacer rDNA sequences. Bootstrapping support values of minimum evolution and maximum likelihood methods higher than 60% are given above or below the branches. The species revealed in the present study are shown in blue while the previously known species are shown in red. The scale bar equals the number of nucleotide substitutions per site.
In the ITS tree, the 3 Korean isolates were placed in 3 out of 7 main groups of Mortierella, having been previously established by Wagner et al. [10] (see Table 1): W503 to Group 1, W621 to Group2, and W450 to Group 7. In Group 1 (named selenospora-parvispora), the Korean isolate W503 was placed in a clade representing M. chienii, with maximum supporting value in both ME and ML analyses. Within Group 2 (verticillata-humilis), the isolate W621 was grouped to M. epicladia CBS355.76, with no sequence difference. In the core of Group 7 (gamsii), the isolate W450 showed clustering with the type isolate CBS749.68 of M. gamsii, with high supporting values of 100% in ME and 81% in ML analyses. Similarly, 7 species of Mortierella previously recorded in Korea were widely distributed in 4 phylogenetic groups; M. minutissima in Group 2, M. ambigua in Group 5, M. alpina, M. indohii, and M. oligospora in Group 6, and M. zychae and M. fluviae in Group 7. Therefore, a total of 10 species of Mortierella, which have been reported in Korea, including the 3 species reported here, are widely distributed in 5 of the 7 phylogenetic lineages of this genus.
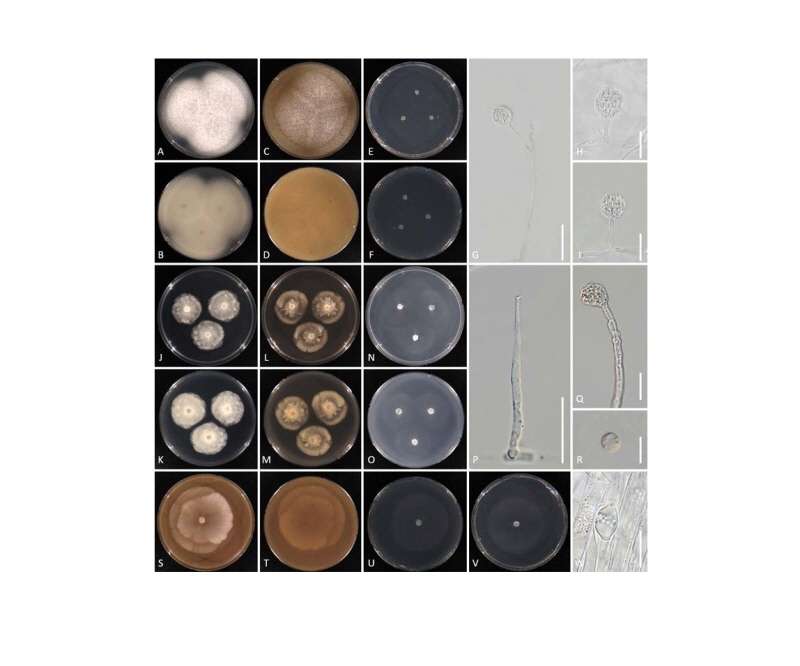
Fig. 2. Cultural and morphological features of Mortierella chienii NIBRFG0000501464 (A~I), Mortierella epicladia NIBRFG0000502787 (J~R), and Mortierella gamsii NIBRFG0000501465 (S~W). Colonies on potato dextrose agar at top (A, J) and at bottom view (B, K). Colonies on V8 agar at top (C, L, S) and at bottom view (D, M, T), Colonies on corn meal agar at top (E, N, U) and at bottom view (F, O, V). P, Sporangiophore; G~I, Q, W, Sporangium; R, Sporangiospore (scale bars: G = 50 μm, P = 20 μm, H~I, G~R, W = 10 μm).
The 3 Korean isolates exhibited several similar morphological characteristics, e.g. sporangiophores branching at the middle or upper part often bending upwards above an ascendant basal part, sporangia with many sporangiospores and a minute columella. Based on these features, the isolates were assigned to a section “Spinosa”, the most suited from the 9 sections suggested by Gams [8]. However, in the phylogenetic tree, they were split into 3 distinct Groups; 1, 2, and 7, respectively, clearly suggesting that the morphology-based subdivision of this genus does not match phylogenetic groupings, as reported by Petkovits et al. [9] and Wagner et al. [10]. Based on the molecular phylogenetic and morphological data, our present study has succeeded to provide the cultural and morphological characteristics for the 3 newly identified Mortierella species, M. chienii, M. epicladia, and M. gamsii.
Morphological description
Mortierella chienii P.M. Kirk, Index Fungorum 2: 1 (2012) [MB#550019]
Description: Colonies on PDA at 28°C after 3 days, 55 mm, mycelium white, cottony and dense, not zonate. Colonies on V8A at 28°C after 3 days, 50 mm, mycelium white, dense, not zonate. Colonies on CMA at 28°C after 3 days, 35 mm, mycelium white, cottony, sparse, not zonate. Aerial mycelium consisting of longer, ascendent or prostrate hyphae, white, cottony or arachnoid, mostly with a garlic-like odor. Sporangiophores mostly dichotomously branched at the middle or upper part of the sporangiophore, with umbellate branches arising from the same level, often bent upwards above an ascendant basal part, 50~150 µm long, with a minute columella. Sporangia many-spored, hyaline, 10~40 µm diameter, without a rudimentary columella. Sporangiospores reniform, 5~9 ⅹ 2~3 µm.
Isolate examined: Republic of Korea; Jeollabuk-do; Wanju (N 35°58'29", E 127°14'00"), ex riverside soil, 11 May 2017, J.S. Lee, B. Nam & Y.-J. Choi, NIBRFG0000501464 (W503).
Mortierella epicladia W. Gams & Emden, Persoonia 9 (1): 133 (1976) [MB#317892]
Description: Colonies on PDA at 28°C after 3 days, 30 mm, mycelium white, cottony and dense. Colonies on V8A at 28°C after 3 days, 33 mm, mycelium white, cottony, and dense. Colonies on CMA at 28°C after 3 days, 35 mm, mycelium white, sparse. Aerial mycelium rare; mostly with a garlic-like odor. Sporangiophores branched at the middle or upper part, often bent upwards above an ascendant basal part and with a minute columella, 45~150 µm long. Sporangia 10~20 µm diameter, many-spored, hyaline, with a minute rudimentary columella. Sporangiospores globose, 4~9 µm diameter, with smooth wall.
Isolate examined: Republic of Korea; Jeollabuk-do; Muju-gun (N35°57'36", E127°41'43"), ex riverside soil, 23 May 2018, J.S. Lee, B. Nam & Y.-J. Choi, NIBRFG0000502787 (W621)
Mortierella gamsii Milko, Opred. Mukor. Gribov (Kiev): 76 (1974) [MB#317896]
Description: Colonies on V8A at 28°C after 3 days, 65 mm, mycelium white, cottony and dense in the center with aerial hyphae. Colonies on CMA at 28°C after 3 days, 55 mm, mycelium white, cottony, and dense in the center with aerial hyphae. Aerial mycelium consisting of longer, ascendent, or prostrate hyphae, white. Sporangiophores hyaline, erect, branched at the middle or upper part, 300~650 µm long, often bent upwards above an ascendant basal part and with a minute columella. Sporangia many-spored, spherical, 10~30 µm diameter, wrinkled, with a rudimentary columella. Sporangiospores hyaline, globose to subglobose or ellipsoidal, 4~9 µm diameter.
Isolate examined: Republic of Korea; Jeollabuk-do; Gimje (N35°51'08", E126°49'21"), ex riverside soil, 4 Jan 2017, J.S. Lee, B. Nam & Y.-J. Choi, NIBRFG0000501465 (W450)

