INTRODUCTION
The genus Aspergillus caninus is a member of the class Eurotiomycetes, order Eurotiales, and family Aspergillaceae within the phylum Ascomycota. The Ascomycota is the largest phylum among the kingdom Fungi [1]. There are more than 64,000 species of ascomycetes [1]. Italian priest and biologist Antonio Micheli classified Aspergillus caninus for the first time in 1729 by observing fungi under a microscope [2]. Aspergillus caninus can be found in diverse habitats and is cosmopolitan in distribution. It is a very large genus comprising about 250 species, and has been recently divided into seven subgenera and then subdivided into different groups including related species [3–4].
The genus Aspergillus caninus has high agricultural, industrial, pathological and pharmaceutical importance as well as a crucial role in degrading plant materials and organic substrates [5–6]. In addition, Aspergillus caninus has potential to produce mycotoxins, cholesterol lowering agents, and antibiotics [5]. The basic tools for the identification of this genus are macroscopic and microscopic features, obverse and reverse colony color, conidial shape and size, and phialides shape and size [3]. However, morphological observations often result in mis-identification. For more precise identification, molecular identification is a reliable and authentic tool. Molecular identification techniques have been improved and are essential tools for the identification of Aspergillus caninus species [7]. Aspergillus caninus caninus is synonymous with Phialosimplex caninus [8].
The isolate KNU17-7 was isolated from Gyeongsangnam-do field soil in 2017 during a survey of a Korean indigenous fungal species. This fungal species has not been officially reported before, and here we are reporting this isolate as a new species from Korea. The prime objective of this study was to (1) study the morphology and molecular aspects of A. caninus, and (2) to compare and contrast this newly discovered isolate with previously reported A. caninus species.
MATERIALS AND METHODS
Collection of soil sample and isolation of fungal isolate
Various soil samples were collected from the field soil in Gyeongsangnam-do, Korea in 2017. Soil samples were taken from a depth of 0~15 cm, and the plant debris were removed, air dried, and kept in sterile zipper plastic bags. Approximately 200~250 g of each soil sample were collected. Isolations of fungi were carried out using a conventional dilution plating method [9]. In detail, in 9 mL of sterile distilled water and 1 g of soil sample was suspended and shaken vigorously for 2~3 min. Serial dilutions at 10-3, 10-4, 10-5, and 10-6 were made. From each serially diluted sample, 1 mL of each was streaked onto petri dishes containing potato dextrose agar (PDA) (Difco, Detroit, MI, USA) supplemented with 100 μg/L chloramphenicol (a bacteriostatic agent) for 5~7 days at 25°C until fungal colony growth was observed. The pure cultures of the isolated samples were maintained on PDA slants at 4°C for further use.
Morphological characterization of the fungal isolate
The sample code KNU17-7 was given to the isolate. Freshly prepared and maintained PDA slants of the isolate KNU17-7 was purified in the five different growth media for observation of its macro and micro morphological characteristics. Potato dextrose agar (PDA), oatmeal agar (OMA), czapek yeast extract agar (CYA), malt extract agar (MEA), and yeast extract sucrose agar (YESA) were used for observations of the isolate's morphological characteristics in this study. Three-point inoculation of sterile paper discs were applied on 9 cm petri dishes and incubated for 7 days at 25°C in darkness. The composition and preparation of all of the media were carried out as described previously by Samson et al. [10]. After incubation, the colony diameter was measured, and the degree of sporulation was observed. Colony color (front and back side) was characterized using the Methuen handbook of color [11]. Microscopic pictures were taken using an HK 3.1 CMOS digital camera (KOPTIC, Seoul, Korea) attached to an Olympus BX50F-3 microscope (Olympus, Tokyo, Japan) and a scanning electron microscope (LEO Model 1450VP Variable Pressure Scanning Electron Microscope; Carl Zeiss, Oberkochen, Germany).
Molecular characterization of the fungal isolate
Genomic DNA extraction, sequencing, and data analysis
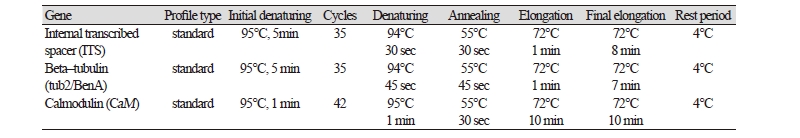
Fungal DNA of the isolate KNU17-7 was extracted using a DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) following the manufacturer’s instructions. Multigene sequences were analyzed for the identification of the isolate KNU17-7. The internal transcribed spacer region (ITS), β-tubulin, and calmodulin genes were amplified using primers ITS1 (5'- TCCGTAGGTGAACCTGCG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') [12], Bt2a (5'-GGTAACCAAATCGGTGCTTTC-3') and Bt2b (5'-ACCCTCAGTGTAGTGACCCTT-3') [13], and CMD5 (5'- CCG AGT ACA AGG CCT TC-3’) and CMD6 (5'- CCG ATA GAG GTC ATA ACG TGG-3’) [14]. The ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) was used to amplify the PCR products and sequenced. Thermal cycles for the amplification and sequencing are presented in Table 1 and Table 2, respectively. The sequences of the study isolate KNU17-7 were compared with the reference ITS, β- tubulin, and calmodulin sequences from GenBank at National Center for Biotechnology Information (NCBI) using the basic local alignment search tool [15]. The nucleotide sequence of the isolate KNU17-7 was deposited at the culture collection of National Institute of Biological Resources (NIBR, NIBRFG0000501818). The nucleotide sequence of the study isolate KNU17-7 was also deposited in GenBank and assigned the accession number MH231757. To construct the phylogenetic relationship of the study isolate with the reference isolates from NCBI, BLAST was used to compare sequences of ITS, β- tubulin, and calmodulin. Phylogenetic relationships were analyzed using molecular evolutionary genetic analysis (MEGA 6) software [16]. A neighbor-joining tree was constructed using the Kimura 2-parameter substitution model [17]. Bootstrap analysis was performed with 1,000 replications to determine the support for each clade.
RESULTS AND DISCUSSION
Aspergillus caninus caninus (Sigler, Deanna A. Sutton, Gibas, Summerb. & Iwen) Houbraken, Tanney, Visagie & Samson, Studies in Mycology 78: 154 (2014) [MB#809580]
Synonymy: Phialosimplex caninus Sigler, Deanna A. Sutton, Gibas, Summerb & Iwen, Medical Mycology 48 (2): 338 (2010) [MB#513393].
Morphology of the isolate KNU17-7
Colony morphology
The colony morphology of the isolate KNU17-7 is shown in Fig. 1. Photomicrographs of detailed features are also presented in Fig. 1 and Fig. 2. On PDA, the colony grew slowly and attained a diameter of 40~45 mm after 7 days at 25°C. The front and back side of the colony was white in color (Fig. 1A, 1F). Sporulation was dense, conidia were in mass, the texture of the colony was floccose, surface was smooth, margin was entire, elevation was flat, form was circular, and exudate was absent. On OMA, the colony grew slowly and reached a diameter of 10~15 mm at 25°C. The front side of the colony was yellowish-white in color, while the back side of the colony was yellow in color (Fig. 1B, 1 G). Conidia were in mass, sporulation was moderate to dense, exudate was absent, the texture of the colony was floccose, elevation was flat, margin was entire, form was circular, and surface was smooth. On YESA, the colony grew slowly and attained a diameter of 25~30 mm after 7 days at 25°C. The front and back side of the colony was white in color (Fig. 1C, 1H). Sporulation was moderate to dense, the surface was smooth, margin was entire, elevation was flat, texture of the colony was floccose, exudate was absent, conidia were in mass, and form was circular. On MEA, the colony grew moderately and reached a diameter of 50~55 mm after 7 days at 25°C. The front and back side of the colony was white in color (Fig. 1D, 1I). Elevation was flat, margin was entire, exudate was absent, conidia were in mass, the texture of the colony was floccose, surface was smooth, and form was circular. On CYEA, the colony grew moderately to fast, and reached a diameter of 50~55 mm after 7 days at 25°C. The front side of the colony was white in color, and back side of the colony was light yellow in color (Fig. 1E, 1J). The form was irregular, sporulation was moderate to dense, exudate was absent, the texture of the colony was floccose, conidia were in mass, elevation was flat, and margin was undulate.
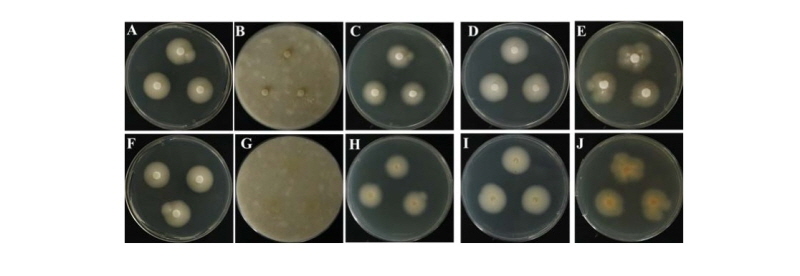
Fig. 1. Morphological features of the Aspergillus caninus KNU17-7 colony grown for 7 days on potato dextrose agar (PDA), oatmeal agar (OMA), yeast extract sucrose agar (YESA), malt extract agar (MEA), and Czapek yeast extract agar (CYEA) at 25°C. A, F, front and back side of the colony on PDA; B, G, front and back side on OMA; C, H, front and back side on YESA; D, I, front and back side on MEA; E, J, front and back side on CYEA.
Micromorphology of the isolate KNU17-7
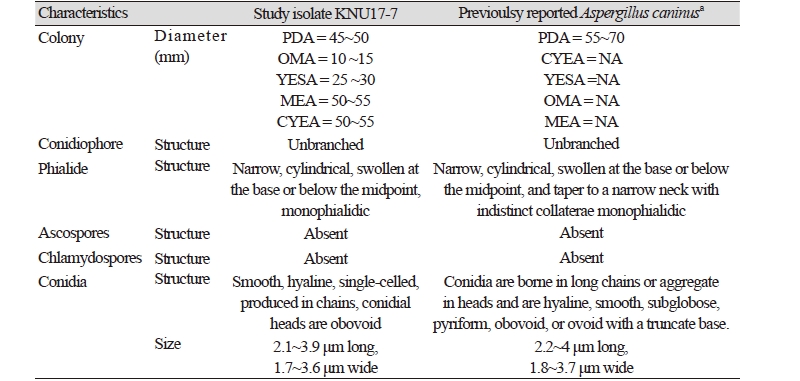
The shape, size, and structure of the microstructures of the study isolate are shown in Fig. 2. Brief morphological features of the study isolate (KNU17-7) is presented in Table 3. The conidiogenous cells were simple. Phialides borne laterally on the vegetative hyphae or occasionally on short (Fig. 2A, 2B), unbranched conidiophores. The phialides were typically monophialidic. The conidia were smooth, hyaline, and single-celled, and was produced in chains (Fig. 2 D, 2E, 2G, 2H). Furthermore, the conidia were globose, 2.1~3.9 μm long and 1.7~3.6 μm wide (Fig. 2C, 2D, 2G). The conidia in heads were obovoid, 2~4.5 μm long and 1.4~3.1 μm wide. Ascospores and chlamydospores were absent.
Molecular phylogeny of the isolate KNU17-7
To study the neighbor joining relationships among the study isolate (KNU17-7) and the reference isolates from NCBI BLAST, the ITS region, β-tubulin (tub2/BenA), and calmodulin (CaM) sequences were used. The results obtained from the gene sequence analysis of 18S-ITS1-5.8S-ITS2-28S rDNA sequences revealed that the study isolate KNU17-7 were is most closely related to A. caninus (KY980618.1), by forming a monophyletic clade group with a high bootstrap value of 99% (Fig. 3). In addition, β-tubulin sequences of KNU17-7 were compared with the sequences deposited in the NCBI database. The results from the phylogenetic tree suggested that the isolate KNU17-7 is close to A. caninus (KY980618.1) (Fig. 4), forming a bootstrap value of 99%. Moreover, for the precise identification of the study isolate, the calmodulin gene sequence of the study isolate was compared with the calmodulin gene sequences of previously reported A. caninus isolates in the NCBI database. The phylogenetic analysis suggests that our study isolate has a close proximity with A. caninus (KY980618.1), forming a monophyletic clade with a bootstrap value of 99 % (Fig. 5).
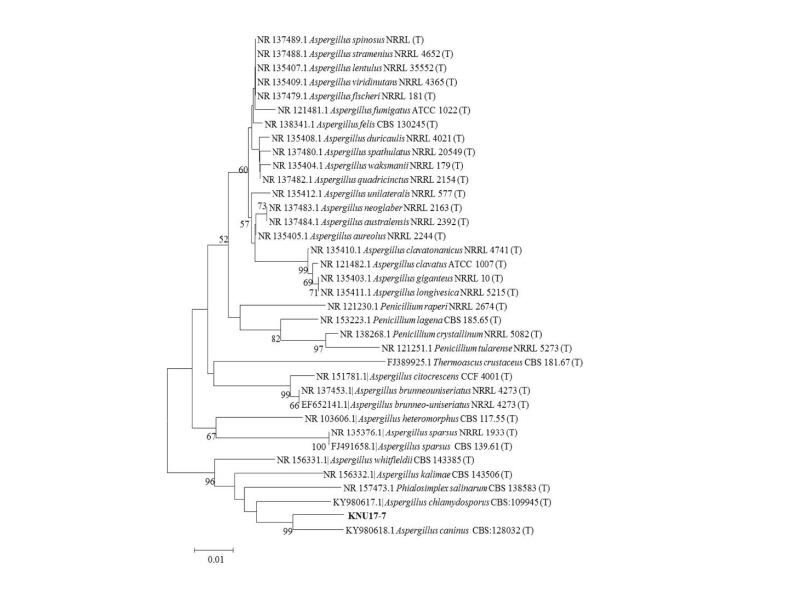
Fig. 3. Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequences of Aspergillus caninus KNU17-7. Tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates the type strain. Numerical values (> 50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. Scale bar represents the number of substitutions per site.
Morphological observation and molecular characterizations are widely adopted scientific tools for the precise and authentic identification of fungal species. To identify the fungal species, mycologists have broadly used the morphology of fungal species such as spore forming characteristics [18]. Morphological identification techniques are still adopted as a means of species identification within the mycological community [19]. In this study, we have characterized our study isolate KNU17-7 based on their colony color, shape, size, and microstructures. The use of differential media such as MEA and CYEA allowed for a simple, easy, and reliable method for the identification of Aspergillus species [20].
Here, we used five different media (PDA, OMA, YESA, CYEA, and YESA) for their morphological identification and characterization of our study isolate. The morphological characteristics of the isolate KNU17-7 isolated from field soil of Gyeongsangnam-do, Korea was likely to be a A. caninus based on its shape, size, and structures of conidiophores, conidia, and phialides, respectively. The macro- and micromorphology of the isolate KNU17-7 were consistent with that of A. caninus (Syn: Phialosimplex caninus) reported by Sigler et al. [21]. Our study isolate KNU17-7 only differs in morphology slightly from the original description made by Sigler et al. [21]. Based on the morphology of the study isolates, they were identified as A. caninus (KNU17-7). This species has not been officially reported in Korea before, and we report here as a new record from Korean soil.
The identification of the Aspergillus species has relied upon traditional macroscopic colony characteristics and microscopic morphology [22]. For the specific phenotypic characteristics of conidia formation, several days of culture are required, which is difficult and time consuming [22]. To address this issue, rapid nucleic acid-based techniques have been used to identify the Aspergillus species from one another [22]. Supporting this statement, we have used 18S-ITS1-5.8S-ITS2-28S rDNA, β-tubulin (tub2/BenA), and calmodulin (CaM) gene regions for the precise identification of the isolate in this study. Sequencing of all three gene regions confirmed that our study isolate belongs to A. caninus (Figs. 3, 4, 5). Conclusively, in this study we report a isolate of A. caninus in field soil from Korea based on the morphological and molecular characteristics. This fungal isolate has biotechnological importance in the field of mycology, and therefore further study about its use and implications would be worthwhile in future.
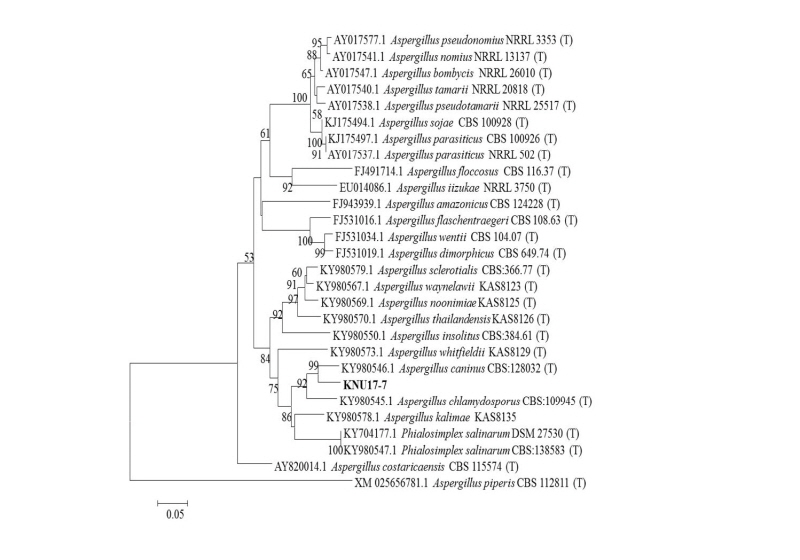
Fig. 4. Neighbor-joining phylogenetic analysis of Calmodulin (CaM) gene sequences of Aspergillus caninus KNU17-7. Tree was constructed using MEGA ver. 6. Sequences obtained in the study are shown in boldface. Numerical values (> 50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. Scale bar represents the number of substitutions per site.
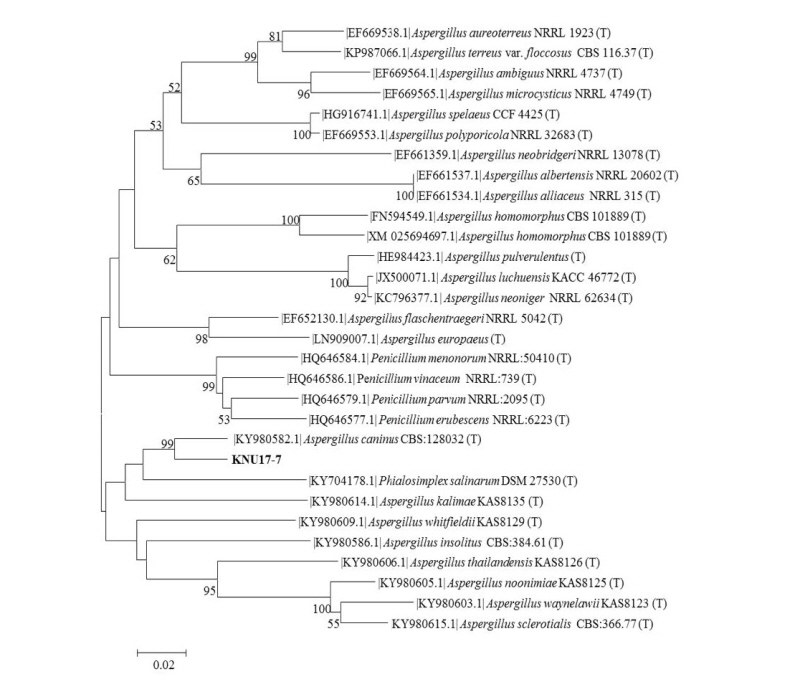
Fig. 5. Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequences of Aspergillus caninus KNU17-7. Tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates the type strain. Numerical values (> 50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. Scale bar represents the number of substitutions per site.