Downy mildew is one of the most common diseases in the world. Members of the family Peronosporaceae (Oomycota), an obligate biotrophic group, are the causal agents of this disease and infect a wide range of mono-and dicotyledonous plants, including many economically important crops [1]. The largest genus of Peronosporaceae, Peronospora, comprises more than 550 validly published taxa [2], which result in high damage on numerous commercially cultivated crops and ornamental plants worldwide [1]. On the family Papaveraceae, about 30 species of Peronospora have been so far described [2, 3]. It also includes downy mildew pathogens of opium poppy (Papaver somniferum), which is a commercially important crop, being cultivated on a large scale for producing seeds and opiums, but also as a valuable ornamental plant in public parks and private gardens. A recent morphological and molecular phylogenetic investigation revealed that two different Peronospora species, P. meconopsidis and P. somniferi, occur on this crop [3]
In Korea, five species of Peronospora have been reported to date, namely, P. arborescens parasitic on opium poppy [4], P. corydalis on Corydalis spp., P. chelidonii on Chelidonium majus var. asiaticum, P. dicentrae on Dicentra spectabilis, and P. hylomeconis on Hylomecon vernale [5, 6]. However, as it has been recently reported that P. arborescens does not cause downy mildew on opium poppy, a further study needs to uncover the identity of the causal agent in Korea. In April 2006, symptoms typical of downy mildew were found on the living leaves of Corydalis ambigua Cham. & Schltdl. (Papaveraceae), growing at the Experimental Forest of Korea University, Yangpyeong, Korea (N37°30'10", E127°43'24"). Corydalis ambigua is a typical spring ephemeral perennial herb that mainly grows in deciduous forests in East Asian countries, including China, Japan, Korea, and Russia [7]. The downy mildew infection resulted in slight discolouration of the leaf tissues and yellow or pale green, vein-limited, polyangular spots on the upper leaf surfaces that developed initially whitish, but then grey growth on the lower surfaces. A representative sample was deposited in the National Institute of Biological Resources (accession no. ZEVCFG0000000023) and the Korea University Herbarium (KUS-F21731).
Detailed morphological characteristics were investigated using an Olympus BX53 microscope (Olympus, Tokyo, Japan), at 100~200ⅹ for conidiophores and 400ⅹ for conidia and ultimate branchlets. DIC micrographs were captured with a DigiRetina 16M camera (Tucsen, Fuzhou, China). The following morphological characteristics were observed. Conidiophores (Fig. 1A~C) emerging through the stomata were tree-like, hyaline, straight to slightly curved, slender, 150~320 μm in height. Trunks were straight or slightly curved, 100~150 μm long, 6~10 μm wide below the first branch, with no callose plug. Basal ends were mostly not differentiated, rarely slightly bulbous, up to 12 μm in width. Branching was mostly monopodial but sometimes trichotomously or subdichotomous, 4~6 orders. Ultimate branchlets (Fig. 1D~G) were mostly in pairs but rarely single, slightly curved to substraight, 4~18 μm long, 1.5~2.5 μm wide at the base, with obtuse tips. Conidia (Fig. 1H~K) were pale olivaceous to olivaceous, subglobose to broadly ellipsoidal, 18~24 μm long, 18~22 μm wide. The length to width ratio of the sporangia was 1.1~1.25 (mean = 1.17; n = 87), with the greatest width mostly at the median. Pedicels were seen as a scar of attachment at the base. Haustoria filling the host cell almost completely, branched, hyphal, filiform. Resting organs (Fig. 1L-N) commonly present and clearly visible as yellow to brown dots on both upper and lower surfaces of infected leaves. Oogonia irregular to broadly ellipsoidal to globose, 44~62 μm diam.; wall wrinkled, yellowish, irregularly 1~2 μm thick. Oospores plerotic, globose, 28~35 μm diam., brownish; wall 2~3 μm thick, smooth; periplasm yellowish to brown.
The morphological investigation revealed that this oomycete pathogen unequivocally belongs to the genus Peronospora, and is well consistent with the known characteristics of Peronospora bulbocapni Beck [8-11]. Three species of Peronospora have been accepted as the causal agents of downy mildew on Corydalis spp., namely, P. bulbocapni, P. corydalis, and P. corydalis-intermediae [2]. However, P. bulbocapni was easily distinguishable from other two species by larger and broader conidia; 18~24 ⅹ 18~22 μm in P. bulbocapni (from the present study), 19~23 ⅹ 15~19 μm in P. corydalis, 17~20 ⅹ 14~17 μm in P. corydalis-intermediae [10]. In addition, the oospores of P. bulbocapni (27~35 μm diam.) were much smaller than those of P. corydalis (37~48 μm) and P. corydalis-intermediae (32~48 μm) [8, 9].
Genomic DNA was extracted from the infected plant tissue of two herbarium specimens using the DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA). PCR amplifications were performed with the primers ITS1-O & LR0 for the internal transcribed spacer (ITS) rDNA [12]. Amplicons were purified and sequenced by a DNA sequencing service (Macrogen, Seoul, Korea) with the primers used for amplification, and the resulting sequences were deposited in GenBank (accession nos. MK212929 for KUS-F21731, MK212930 for KUS-F21089). The sequences were aligned using MAFFT 7 [13] with the Q-INS-1 algorithm. Minimum evolution (ME) and maximum likelihood (ML) methods were used to construct phylogenetic trees. ME analysis was done using MEGA 7.0 [14], with the default settings of the program, except for replacement with the Tamura-Nei model. For ML analysis, 1,000 rounds of random addition of sequences and 1,000 fast bootstrap replicates were performed with RAxML 7.0.3 [15] using the GTRCAT model. The Korean isolate from Corydalis ambigua exhibited only two nucleotide differences out of 800 characters with P. bulbocapni originated from C. cava (AY198272) for the ITS sequences. The slight sequence differences between the Korean samples and P. bulbocapni s. s. may be due to either the different host species (C. ambigua vs C. cava) or the distant geographic origins (Korea vs Europe), as in this study no morphological differences were found between them (data not shown). As the ME and ML trees were congruent, only a ME tree is shown in Fig. 2. In the ITS tree, the Korean samples were grouped with P. bulbocapni with high supporting values of 98% in ME and 100% in ML analyses, supporting the Blastn-based species identification. A further study with additional collections and phylogenetic markers [16] needs to resolve this divergent group, but also to recover the macro-evolutionary relationship between Peronopsora species and their host plants of Coryalis, as has been studied in other downy mildew species [12, 17].
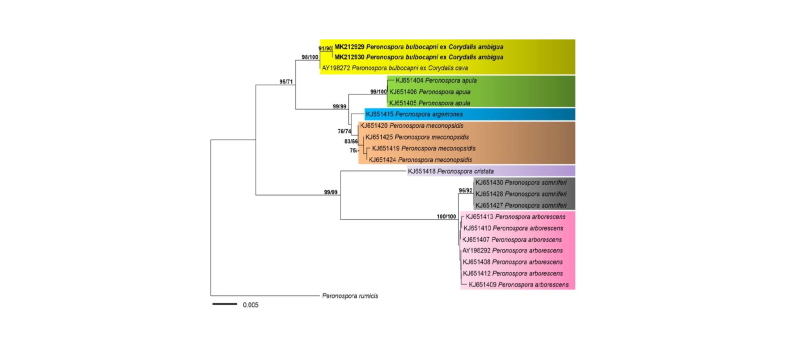
Fig. 2. Minimum evolution tree of Peronospora species using the internal transcribed spacer rDNA sequences. Bootstrapping values (minimum evolution BP/ maximum likelihood BP) higher than 70% are shown above the branches (1,000 replicates). The scale bar equals the number of nucleotide substitutions per site
Based on the morphological and phylogenetic analyses, the downy mildew pathogen on C. ambigua was identified as Peronospora bulbocapni. Until now, about 20 species of Corydalis have been recorded as host plants of downy mildews, among which 6 species were affected by P. bulbocapni [18]. On Corydalis ambigua, infection by Peronospora has been reported only once in Japan in 1931 [18], without any detailed description. The present study confirmed that Corydalis ambigua is a rare host plant of P. bulbocapni in East Asia. To our knowledge, this is the first report of P. bulbocapni occurring on C. ambigua in Korea.


