Introduction
Freshwater fungi, a poly-phylogenetic group, are taxonomically diverse organisms colonizing the substrata in aquatic or semi-aquatic environments [1]. Natural habitats of freshwater fungi include soil, submerged wood and leaves, and endophytes of plants and animals. Recent studies on different habitats of freshwater and marine environments indicated that aquatic fungi can be abundant in eukaryotes depending on the ecosystems and environmental settings [2]. The main role of freshwater fungi is the degradation of dead plant material in water. They may also be involved in the degradation of animal tissue such as insect exoskeletons, fish scales, and hair. Some of these fungi function as plant pathogens or endophytes living on plant tissues [3]. Freshwater fungi have been found to grow on diverse substrates; however, their roles in aquatic ecosystems, and their physiological and biochemical characteristics remain to be determined.
Pezizomycotina is the largest subphylum of Ascomycota, including filamentous ascomycetes. More than 32,000 species belong to this subphylum [4]. Pezizomycotina fungi form varied ecological relationships such as symbiotic (with mycorrhizae, endophytes, and lichens), necrotic (decaying of wood and litter), and pathogenic (for animals and plants) [5]. They are found worldwide and in various habitats including aquatic and terrestrial environments. Many Ascomycota fungi are found in freshwater environments [6].
For the first time, we isolated eleven Pezizomycotina species (Cladosporium angulosum, Pseudorobillarda phragmitis, Paraconiothyrium estuarinum, Pseudopithomyces palmicola, Pyrenochaetopsis paucisetosa, Thelebolus globosus, Plagiostoma mejianum, Trichoderma cremeum, Fusarium tanahbumbuense, Coniochaeta endophytica, and Chaetomium tenue) from environmental samples such as plant litter, sediment, and filtered water in Korean rivers, wetland, and streams, and their molecular phylogenetic and morphological characteristics were investigated.
MATERIALS AND METHODS
Isolation of fungal strains and culture conditions
Fungal strains were collected from plant litter, sediment, and filtered water sampled from freshwater streams and rivers. The collection information of all strains identified in this study was listed in Table 1. To isolate fungal strains, plant litter samples were washed with distilled water at least twice, and incubated in a pretreatment liquid medium (0.05% 3-morpholinopropane-1-sulfonic acid [weight/volume (w/v)], 0.05% KNO3 [w/v], 0.025% KH2PO4 [w/v], and 0.025% K2HPO4 [w/v]) at 20℃ for three days. Then, 100 µL of the pretreatment medium was spread on a 1% water agar plate and incubated at 20℃ for two days. Hyphal tips and germinated conidia were isolated under a microscope and transferred onto a 24-well plate containing V8 agar (V8A; 8% V8 juice [v/v] and 1.5% agar [w/v], adjusted to pH 6.0 using 10 N NaOH) and incubated at 25℃ in the dark. To isolate fungal strains from sediments, a dilution method was used. Diluted suspension (200 μL) of freshwater sediment and distilled water (1:200 and 1:2,000) were spread on potato dextrose agar (PDA; 3.9% potato dextrose agar powder [w/v]; Difco, Sparks, MD, USA) containing 50 ppm streptomycin, and fungal strains were isolated in the pure form after incubation for 4-5 days at 25℃ by repeating this step. To isolate fungal strains directly from water, 50 mL of water was filtered through a membrane filter (Merck Milliphore, Cork, Ireland), and membranes were laid on to the surface of the agar plates (2% agar [w/v], supplemented with 50 ppm of streptomycin) and incubated overnight at 15℃. After 1 day, the membrane was removed, and hyphal tips and germinated conidia were isolated under a microscope and transferred to a new medium. Fungal strains were isolated in the pure form after incubation for 4-5 days at 25℃ by repeating this step. All strains identified in this study were grown on malt extract agar (MEA; 2% malt extract [w/v] and 2% agar [w/v]), oatmeal agar (OA; 7.25% oatmeal agar powder [w/v]; Difco, Sparks, MD, USA), potato carrot agar (PCA; 2.4% potato carrot agar powder [w/v]; HiMedia, Mumbai, India), synthetic nitrogen-poor or nutrient-poor agar (SNA; 0.02% sucrose [w/v], 0.02% glucose [w/v], 0.1% KNO3 [w/v], 0.1% KH2PO4 [w/v], 0.05% MgSO4·7H2O [w/v], 0.05% NaCl [w/v] and 1.2% agar [w/v]) and corn meal dextrose agar (CMDA; 2% cornmeal [w/v], 2% glucose [w/v], and 2% agar), and yeast extract peptone dextrose agar (Duchefa Biochemie, Haarlem, the Netherlands).
Morphological analysis
Microstructures of fungal species were observed under an Eclipse Ni light microscope (Nikon, Tokyo, Japan) equipped with a Ds-Ri2 digital camera (Nikon, Tokyo, Japan). At least 50 individuals were examined for observation and measurement of each structure.
DNA extraction, polymerase chain reaction (PCR), and DNA sequencing
Fungal genomic DNA was isolated using a NucleoSpin® Plant II DNA extraction kit (Macherey-Nagal, Düren, Germany). For molecular identification of the fungi, PCR amplifications were performed for the internal transcribed spacer (ITS) rDNA region using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [7], for the large subunit of rDNA (LSU) using primers LR0R (5′-ACCCGCTGAACTTAAGC-3’) and LR7 (5′-TACTACCACCAAGATCT-3′) [8], for the beta-tubulin gene (TUB) using primers bt2a (5′-GGTAACCAAATCGGTGCTGCTTTC-3′) and bt2b (5′-ACCCTCAGTGTAGTGACCCTTGGC-3′) [9], for the translation elongation factor 1 gene (EF1) using primers EF1-983F (5′-GCYCCYGGHCAYCGTGAYTTYAT-3′) and EF1-1576R (5′-ACHGTRCCRATACCACCRATCTT-3′) [10], for the RNA polymerase II gene (RPB2) using primers fRPB2-5F (5′-GAYGAYMGWGATCAYTTYGG-3′) and fRPB2-7cR (5′-CCCATRGCTTGYTTRCCCAT-3′) [11], and for the actin gene (ACT1) using primers ACT-512F (5’- ATGTGCAAGGCCGGTTTCGC-3’) and ACT-783R (5’-TACGAGTCCTTCTGGCCCAT-3’) [12]. Amplicons were sequenced by a DNA sequencing service (Macrogen Inc., Daejoen, Korea) using the same primers as those used for amplification. A homology search of the DNA sequences was performed using the BLAST algorithms available from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov).
Phylogenetic analysis
We obtained the sequences of the reference species from NCBI for phylogenetic analyses, which were shown in Figs. 1-11. All reference sequences have been reported previously [13-27]. Sequences were edited using the DNAStar software package version 5.05 (DNAStar, Inc., Madison, WI, USA). The accession numbers of the sequences used in this study were shown in the phylogenetic trees (Figs. 1-11) constructed using the maximum likelihood (ML) method. ML analysis was performed using MEGA 7.1 [28] with the default settings of the program, except for replacement with the Tamura-Nei model. Bootstrapping analysis of 1,000 replicates was performed to test the robustness of each grouping.
RESULTS AND DISCUSSION
Phylogenetic analysis
Phylogenetic analysis was performed to identify and infer the phylogenetic relationships of the eleven Pezizomycotina fungi with other similar species. For the genus Cladosporium and its related species, the ITS sequences were used for phylogenetic analysis. It was evident from Fig. 1I that NNIBRFG27319 formed to a clade with Cladosporium angulosum. In BLASTn analysis, the act1 of NNIBRFG27319 showed a high similarity (96.05%) with that of the C. angulosum strain CPC14566. For Pseudorobillarda species, the ITS sequences were used for phylogenetic analysis. Fig. 2F shows that NNIBRFG6455 formed a clade with two isolates of Pseudorobillarda phragmitis. In BLASTn analysis, the ITS of NNIBRFG6455 showed high similarity (100%) with that of P. phragmitis strain CBS 143381. For the genus Paraconiothyrium and its related species, the TUB sequences were used for phylogenetic analysis. Fig. 3G shows that NNIBRFG27315 formed a clade with Paraconiothyrium estuarinum. In BLASTn analysis, the TUB gene of NNIBRFG27315 showed high similarity (98.47%) with that of the P. estuarinum strain CBS 109850. For Pseudopithomyces genus and its related species, a combination of ITS and LSU sequences was used for phylogenetic analysis. Fig. 4F shows that NNIBRFG26612 formed a clade with three isolates of Pseudopithomyces palmicola including the type material. In BLASTn analysis, the ITS and LSU of NNIBRFG6612 showed high similarity (100% and 99.83%, respectively) with those of P. palmicola. For Pyrenochaetopsis species, a combination of ITS and LSU sequences was used for phylogenetic analysis. Fig. 5I shows that NNIBRFG27317 formed a clade with the type material of Pyrenochaetopsis paucisetosa. In BLASTn analysis, the ITS and LSU of NNIBRFG27317 showed high similarity (100% in both cases) with those of P. paucisetosa. For Thelebolus species, the TUB sequences were used for phylogenetic analysis. Fig. 6I shows that NNIBRFG5834 formed a clade with the type material of Thelebolus globosus. In BLASTn analysis, the TUB gene of NNIBRFG5834 showed high similarity (95.08%) with that of T. globosus. For Plagiostoma species and its related species, the TUB sequences were used for phylogenetic analysis. Fig. 7G shows that NNIBRFG22701 formed a clade with Plagiostoma mejianum. In BLASTn analysis, NNIBRFG22701 showed high similarity (99.76%) with that of P. mejianum. For Trichoderma species, the RPB2 sequences were used for phylogenetic analysis. Fig. 8I shows that NNIBRFG22286 formed a clade with two isolates of Trichoderma cremeum including the type material. In BLASTn analysis, the RPB2 gene of NNIBRFG22286 showed high similarity (99.66%) with that of T. cremeum. For Fusarium species and related species, the RPB2 sequences were used for phylogenetic analysis. Fig. 9H shows that NNIBRFG27321 formed a clade with the type material of Fusarium tanahbumbuense. In BLASTn analysis, the RPB2 gene of NNIBRFG27321 showed high similarity with of F. tanahbumbuense (99.78%). For Coniochaeta species and related species, the EF1 sequences were used for phylogenetic analysis. Fig.10I showed that NNIBRFG15250 formed a clade with the two isolates of Coniochaeta endophytica. In BLASTn analysis, the EF1 sequences of NNIBRFG15250 showed high similarity (100%) with C. endophytica. For Chaetomium species and related species, the TUB sequences were used for phylogenetic analysis. Fig. 11G showed that NNIBRFG22929 formed a clade with Chaetomium tenue. In BLASTn analysis, the TUB sequences of NNIBRFG22929 showed high similarity (99.77%) with C. tenue.
Taxonomy
Based on the molecular phylogeny and morphological data, we identified eleven fungal species that have not previously been recorded in Korea: Cladosporium angulosum, Pseudorobillarda phragmitis, Paraconiothyrium estuarinum, Pseudopithomyces palmicola, Pyrenochaetopsis paucisetosa, Thelebolus globosus, Plagiostoma mejianum, Trichoderma cremeum, Fusarium tanahbumbuense, Coniochaeta endophytica, Chaetomium tenue.
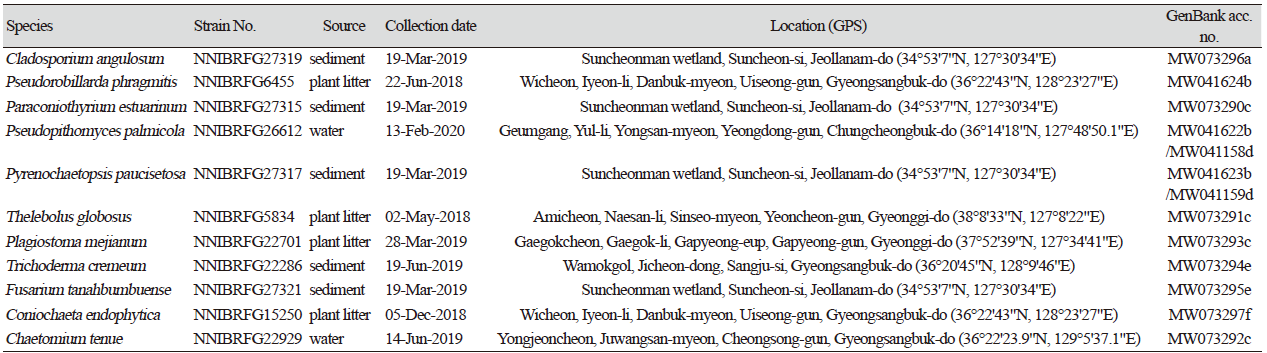
Cladosporium angulosum Sand.-Den., Deanna A. Sutton & Guarro, Persoonia 36: 289 (2016) [MB#815333] (Fig. 1)
Description: Colonies grew quite slowly and reached a diameter of 28 mm on PDA, 31 mm on MEA, 27 mm on OA, 30 mm on PCA, 33 mm on CMDA, and 30 mm on V8A at 25℃, 7 days after inoculation. The color of the colony was white at the margin, turning to gray green, on PDA; white to light gray on OA; whitish with cottony aerial mycelium, turning to light gray on PCA; dark green with floccose aerial mycelium on MEA; translucent at margin to dark green at center on CMDA; and translucent white to light gray with fluffy aerial mycelium on V8A. One-celled conidia were formed as densely branched chains, and subglobose or obovoid, measuring 3.3-6.9 µm×1.9-3.7 μm (x=4.79±0.71 µm×2.85±0.38 μm, n=50, length/width [L/W] ratio=1.68).
Habitat: Sediment in wetland
Specimen examined: Suncheonman wetland, Suncheon-si, Jeollanam-do, Republic of Korea, 19 Oct 2019, NNIBRFG27319, Nakdonggang National Institute of Biological Resources
Note: The type material of C. angulosum, CPC 22271, was isolated from indoor air sample, and other strains were collected from plant [13]. In the present study, NNIBRFG27319 was isolated from wetland sediment.
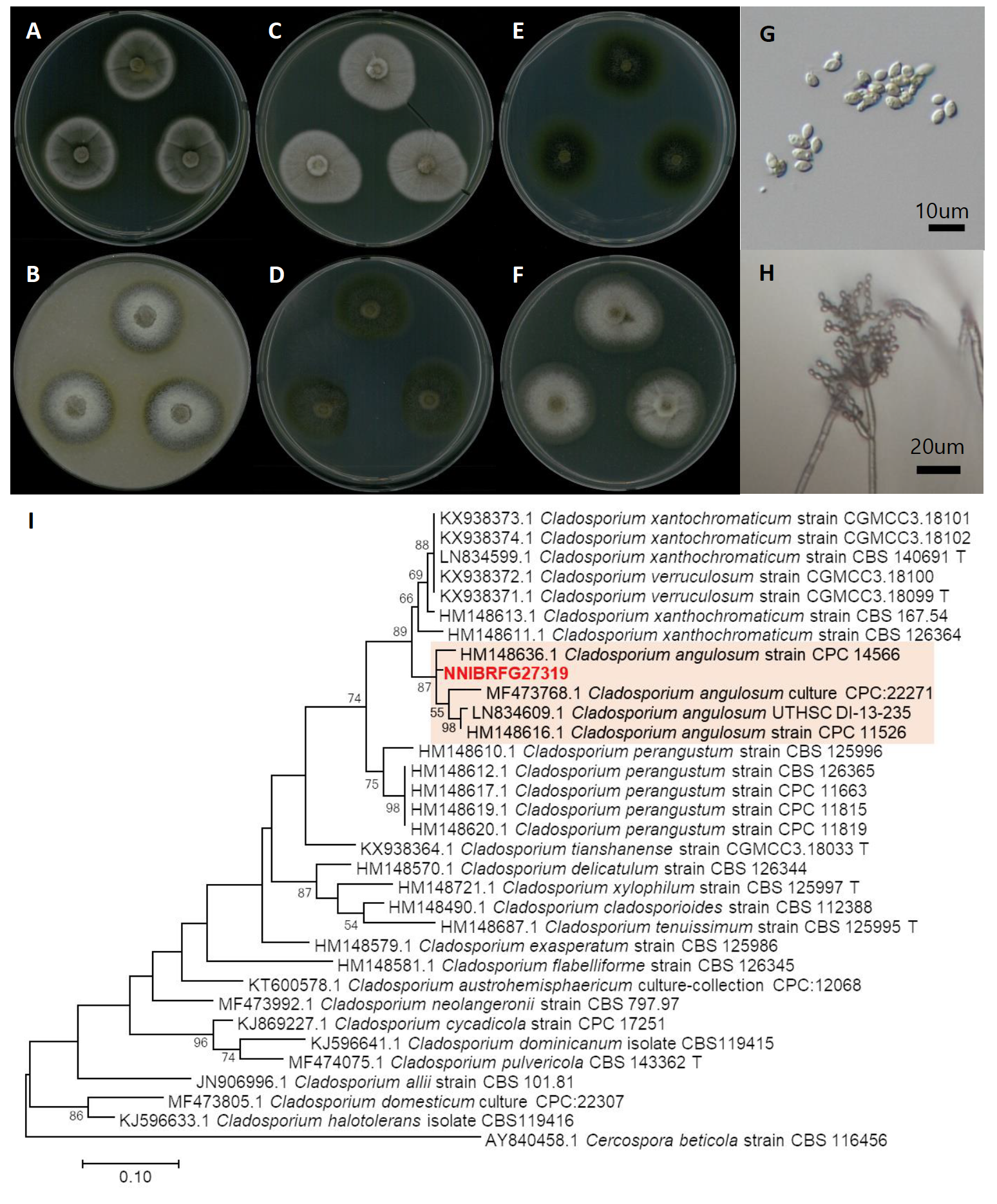
Fig. 1.Characters of Cladosporium angulosum NNIBRFG27319. Mycelial growth on (A) potato dextrose agar (PDA), (B) oatmeal agar (OA), (C) potato carrot agar (PCA), (D) malt extract agar (MEA), (E) corn meal dextrose agar (CMDA), (F)V 8-juice agar (V8A) for 7 days at 25℃. Microscopic observation showed (G) conidia and (H) conidiphore (Scale bars: 10 μm). Phylogenetic tree of Cladosporium angulosum NNIBRFG27319 and related species, based on maximum-likelihood analysis of (I) the actin gene sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.
Pseudorobillarda phragmitis (Cunnell) M. Morelet, Bulletin de la Société des Sciences Naturelles et d'Archéologie de Toulon et du Var 175: 5 (1968) [MB#321865] (Fig. 2)
Description: Colonies grew slowly and reached a diameter of 18 mm on MEA, 17 mm on OA, 24 mm on CMDA, 16 mm on YPDA at 25℃, 7 days after inoculation. The color of the colony was hyaline with a smooth surface on MEA and CMDA; white with fluffy dense aerial mycelium on YPDA; and white to light gray with dense aerial mycelium on OA. Conidia were hyaline, fusiform, straight, one-septate, bearing 2-5, divergent, apical appendages (10-20 µm long, x=11.49±1.97μm). The size of conidia was 3.3-6.9 µm×1.9-3.7 μm (x=16.33±1.76 µm×3.73±0.76 μm, L/W ratio=4.38, n=50).
Habitat: Plant litter in stream
Specimen examined: Wicheon, Iyeon-li, Danbuk-myeon, Uiseong-gun, Gyeongsangbuk-do, Republic of Korea, 22 Jun 2018, NNIBRFG6455, Nakdonggang National Institute of Biological Resources
Note: The type material of P. phragmitis, IMI 70768, was isolated from dead stems of Phragmites comunis [14,15]. In the present study, NNIBRFG27319 was isolated from plant litter in a stream.
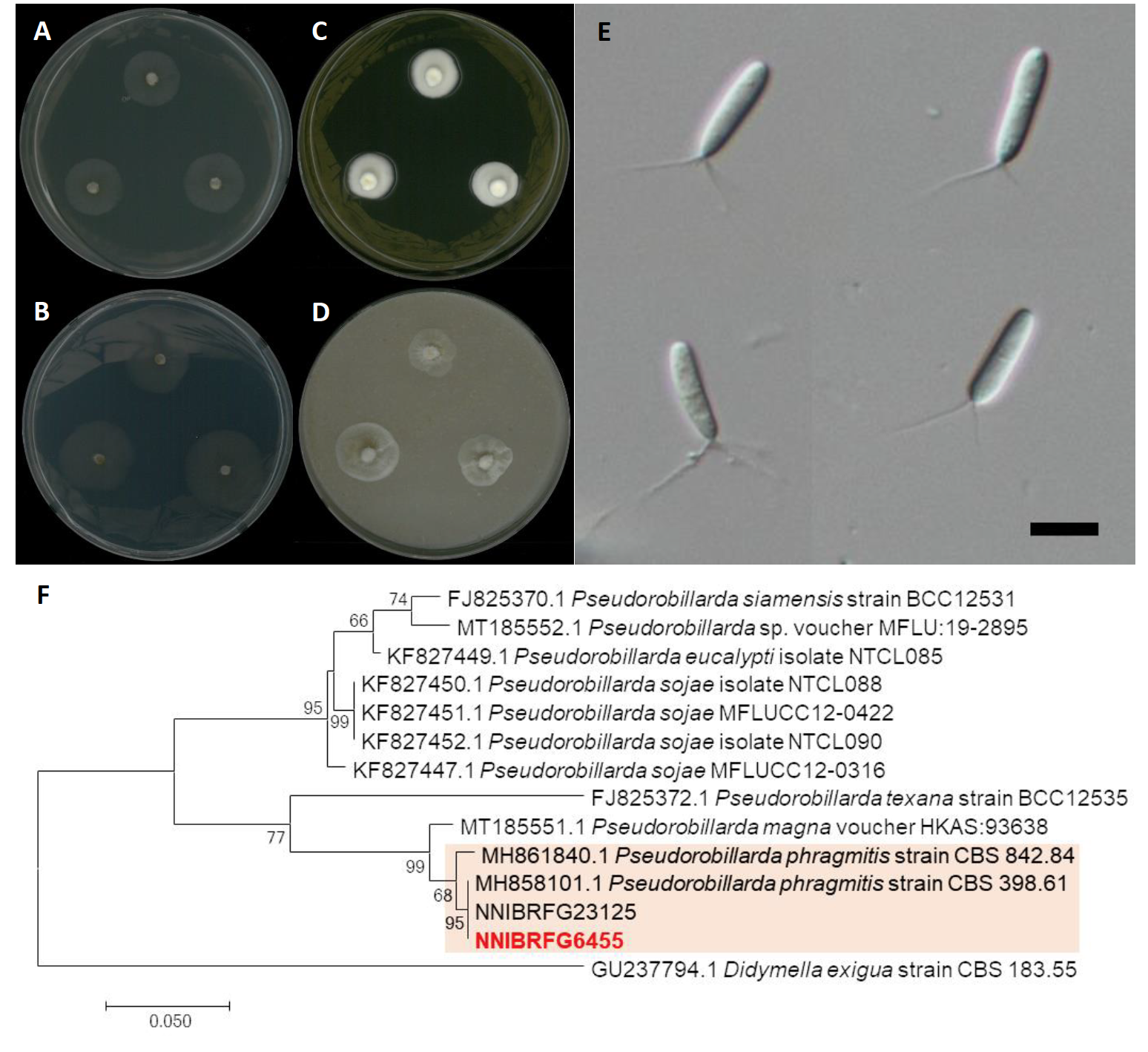
Fig. 2.Characters of Pseudorobillarda phragmitis NNIBRFG6455. Mycelial growth on (A) malt extract agar (MEA), (B) corn meal dextrose agar (CMDA), (C) yeast peptone dextrose agar (YPDA), (D) oatmeal agar (OA) for 7 days at 25℃. Microscopic observation showed (E) conidia (Scale bars: 10 μm). Phylogenetic tree of Pseudorobillarda phragmitis NNIBRFG6455 and related species, based on maximum-likelihood analysis of (F) the internal transcribed spacer (ITS) rDNA sequence. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold.
Paraconiothyrium estuarinum Verkley & M. da Silva, Studies in Mycology 50 (2): 327 (2004) [MB#500081] (Fig. 3)
Description: Colonies grew quite fast and reached a diameter of 44 mm on PDA, 49 mm on CMDA, 49 mm on OA, and 55 mm on V8A at 25℃, 7 days after inoculation. The color of the colony was whitish gray with floccose aerial mycelium, turning to olive-green from center on PDA; hyaline white with short wooly aerial mycelium on CMDA; olivaceous-grey with fluffy aerial mycelium on OA; and white to light gray with cottony aerial mycelium on V8A. Conidia were ellipsoidal or short-cylindrical, rounded at both ends, hyaline, one-celled, and measuring 2.0-4.9 µm×1.3-2.4 μm (x=3.69±0.49 µm×1.83±0.26 μm, L/W ratio=2.00, n=50).
Habitat: Sediment in wetland
Specimen examined: Suncheonman wetland, Suncheon-si, Jeollanam-do, Republic of Korea, 19 Oct 2019, NNIBRFG27315, Nakdonggang National Institute of Biological Resources
Note: The type material of P. estuarinum, CBS 10985, was isolated from sediment of an estuarine [16]. Similarly, in this study NNIBRFG27319 was isolated from wetland sediment.
Fig. 3.Characters of Paraconiothyrium estuarinum NNIBRFG27315. Mycelial growth on (A) potato dextrose agar (PDA), (B) corn meal dextrose agar (CMDA), (C) oatmeal agar (OA), (D) V8-juice agar (V8A) for 7 days at 25℃. Microscopic observation showed (E, F) conidia (Scale bars: 10 μm). Phylogenetic tree of Paraconiothyrium estuarinum NNIBRFG27315 and related species, based on maximum-likelihood analysis of (G) the beta-tubulin sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold.
Pseudopithomyces palmicola J.F. Li, Ariyaw. & K.D. Hyde ex M. Lorenzini & Zapparoli, Sydowia 70: 270 (2018) [MB#825124] (Fig. 4)
Description: Colonies grew slightly fast and reached a diameter of 49 mm on PDA, and 58 mm on OA at 25℃, 7 days after inoculation. The color of the colony was yellowish white at margin, turning to olivaceous brown with dense aerial mycelium on PDA; and whitish gray with short wooly-floccose aerial mycelium, turning to light gray on OA. Conidia were muriform, subglobose, 4-8-septate, echinulate, ovoid, thin-walled, light brown to dark brown, and measuring 16.4-30.3 µm×8.9-18.48 μm (x=24.41±3.41 µm×14.07±2.21 μm, L/W ratio=1.73, n=50).
Habitat: Freshwater
Specimen examined: Geumgang, Yul-li, Yongsan-myeon, Yeongdong-gun, Chungcheongbuk-do, 13 Feb 2020, NNIBRFG26612, Nakdonggang National Institute of Biological Resources
Note: The type material of P. palmicola, MFLU 15-1474, was isolated from leaves of Acoelorrhaphe wrightii [18]. In this study, NNIBRFG26612 was isolated from filtered freshwater in river.
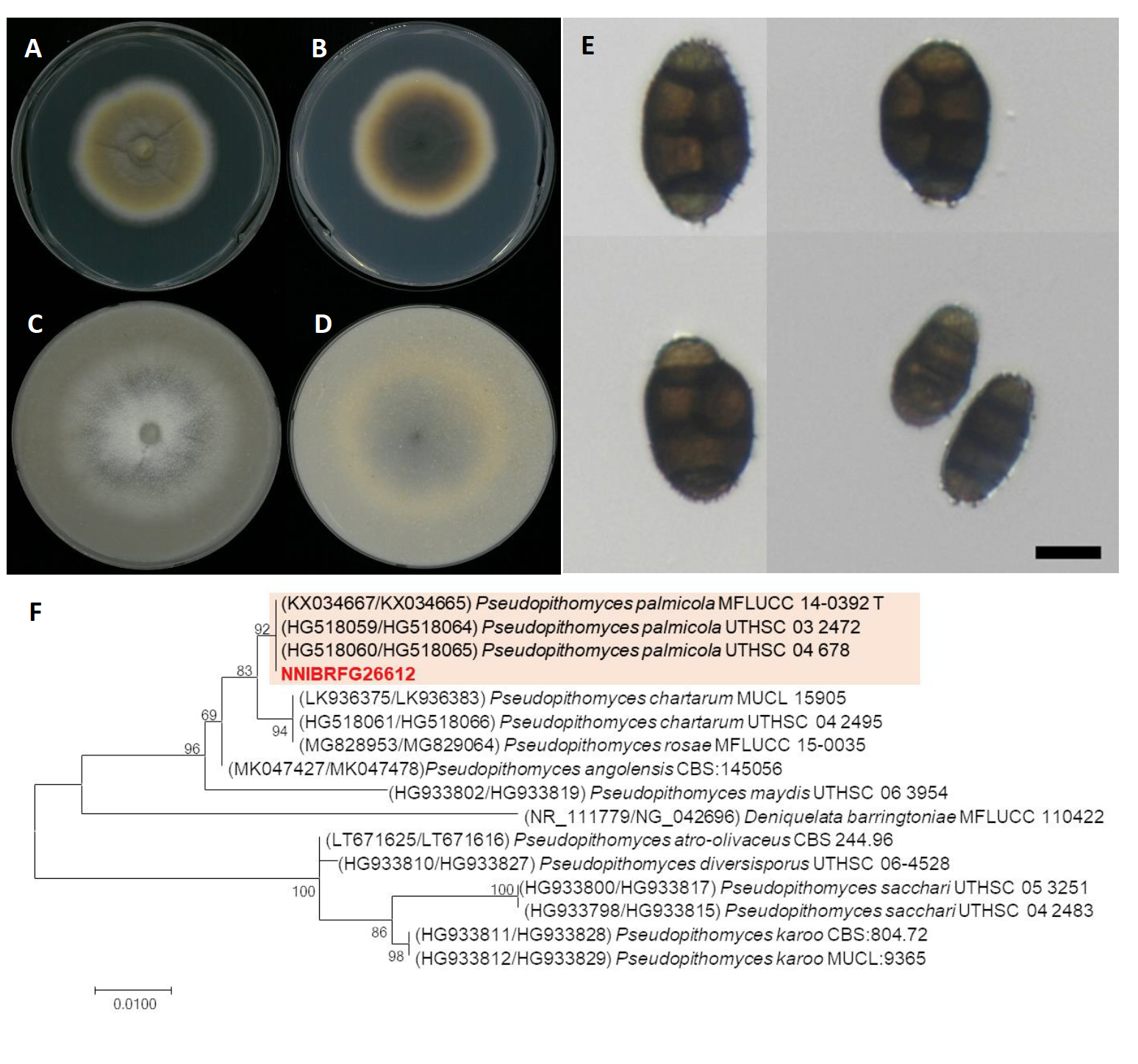
Fig. 4.Characters of Pseudopithomyces palmicola NNIBRFG26612. Mycelial growth on potato dextrose agar (PDA; A, front; B, back), oatmeal agar (OA; C, front; D, back) for 7 days at 25℃. Microscopic observation showed conidia (E)(Scale bars: 10 μm). Phylogenetic tree of Pseudopithomyces palmicola NNIBRFG26612 and related species, based on maximum-likelihood analysis of a combination of internal transcribed spacer (ITS) rDNA and 28S rDNA sequences (F). Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.
Pyrenochaetopsis paucisetosa Valenz.-Lopez, J.F. Cano, Guarro & Stchigel, Studies in Mycology 90: 60 (2017) [MB#819766] (Fig. 5)
Description: Colonies grew slowly and reached a diameter of 15 mm on PDA, 25 mm on OA, 17 mm on PCA, 19 mm on MEA, 24 mm on V8A, and 18 mm on YPDA at 25℃, 7 days after inoculation. The color of the colony was whitish gray with flatted aerial mycelium, turning to light gray on OA; light gray with cottony aerial mycelium on PCA; pale gray with dense aerial mycelium on PDA; yellowish gray with fluffy aerial mycelium on YPDA; light gray with an uneven margin on MEA; gray with floccose aerial mycelium on V8A. Conidia were aseptate, hyaline, cylindrical, and measuring 2.8-4.4 µm×1.4-2.6 μm (x=3.62±0.38 µm×1.94±0.23 μm, L/W ratio=1.86, n=50).
Habitat: Sediment in wetland
Specimen examined: Suncheonman wetland, Suncheon-si, Jeollanam-do, Republic of Korea, 19 Oct 2019, NNIBRFG27317, Nakdonggang National Institute of Biological Resources
Note: The type material of P. paucisetosa, CBS 142460, was isolated from a specimen of superficial human tissue [19]. In this study, NNIBRFG27317 was isolated from sediment in freshwater.
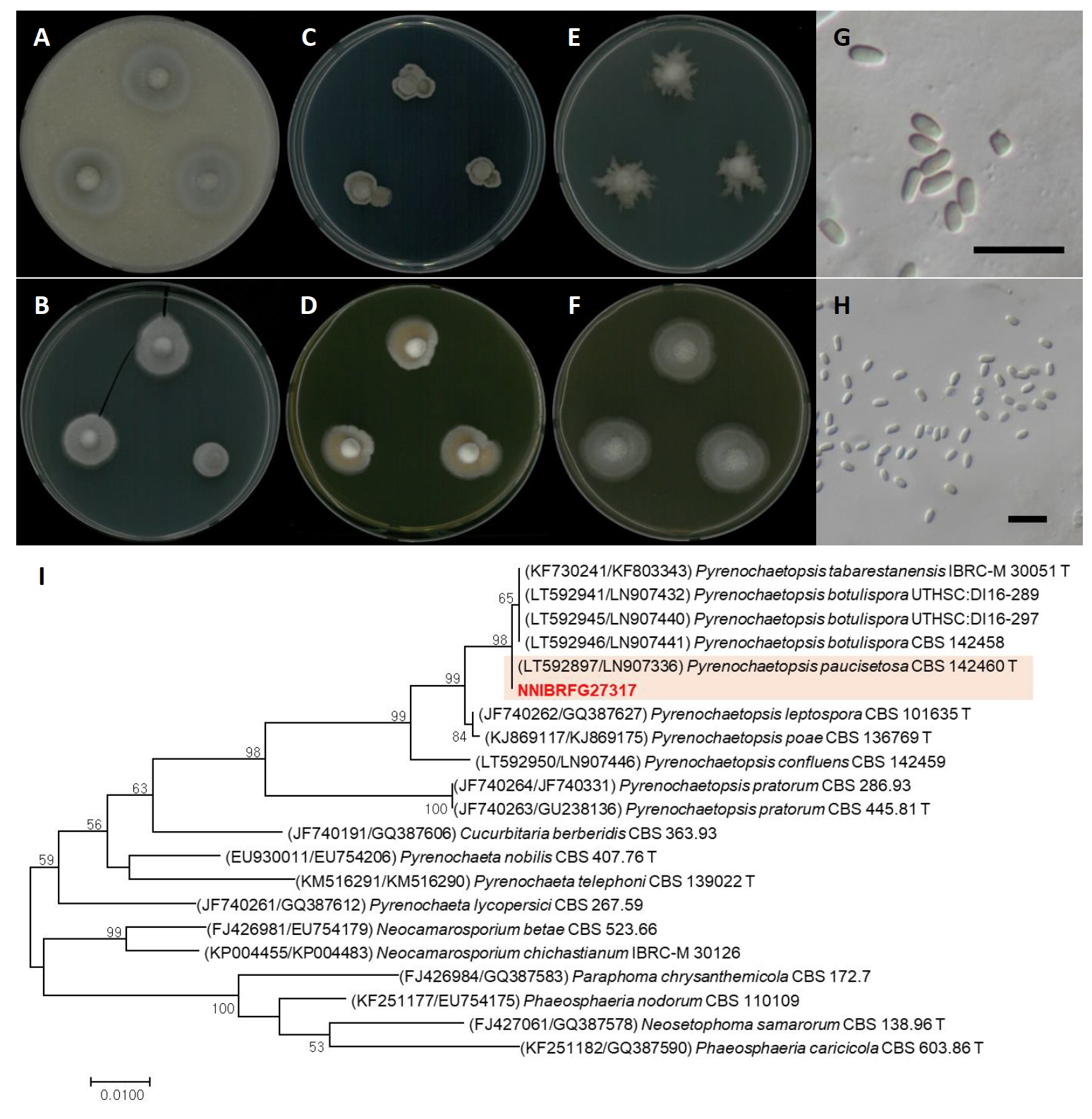
Fig. 5.Characters of Pyrenochaetopsis paucisetosa NNIBRFG27317. Mycelial growth on (A) oatmeal agar (OA), (B) potato carrot agar (PCA), (C) potato dextrose agar (PDA), (D) yeast peptone dextrose agar (YPDA), (E) malt extract agar (MEA), (F) V8-juice agar (V8A) for 7 days at 25℃. Microscopic observation showed (G-H) conidia morphology (scale bars-10 μm). Phylogenetic tree of Pyrenochaetopsis paucisetosa NNIBRFG27317 and related species, based on maximum-likelihood analysis of a combination of (I) internal transcribed spacer (ITS) rDNA and 28S rDNA sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.
Thelebolus globosus Brumm. & de Hoog, Studies in Mycology 51: 72 (2005) [MB#500195] (Fig. 6)
Description: Colonies grew slowly and reached a diameter of 22 mm on PDA, 17 mm on OA, 24 mm on MEA, 30 mm on V8A, 18 mm on CMDA, and 30 mm on YPDA at 25℃, 7 days after inoculation. The color of the colony was hyaline white with smooth aerial mycelium on OA; translucent white to creamish with smooth surface on PDA; hyaline white with smooth aerial mycelium on MEA; translucent white to creamish with a smooth surface on V8A; hyaline white with smooth aerial mycelium on CMDA; yellowish white with a dense smooth surface on YPDA. Conidia were subglobose or very shortly ellipsoid, hyaline, thickwalled, with contents rich in small globular granules, and measuring 2.9-7.7 µm×2.1-5.8 μm (x=5.28±1.08 µm×4.03±0.82 μm, L/W ratio=1.30, n=50)
.Habitat: Plant litter in freshwater
Specimen examined: Amicheon, Naesan-li, Sinseo-myeon, Yeoncheon-gun, Gyeonggi-do, 02 May 2018, NNIBRFG5834, Nakdonggang National Institute of Biological Resources
Note: The type material of T. globosus, CBS 113940, was isolated from biomat in lakes of the Antarctica region [20], and limited geographical distribution of this species has been reported. In this study, NNIBRFG5834 was isolated from plant litter in freshwater in a temperate zone.
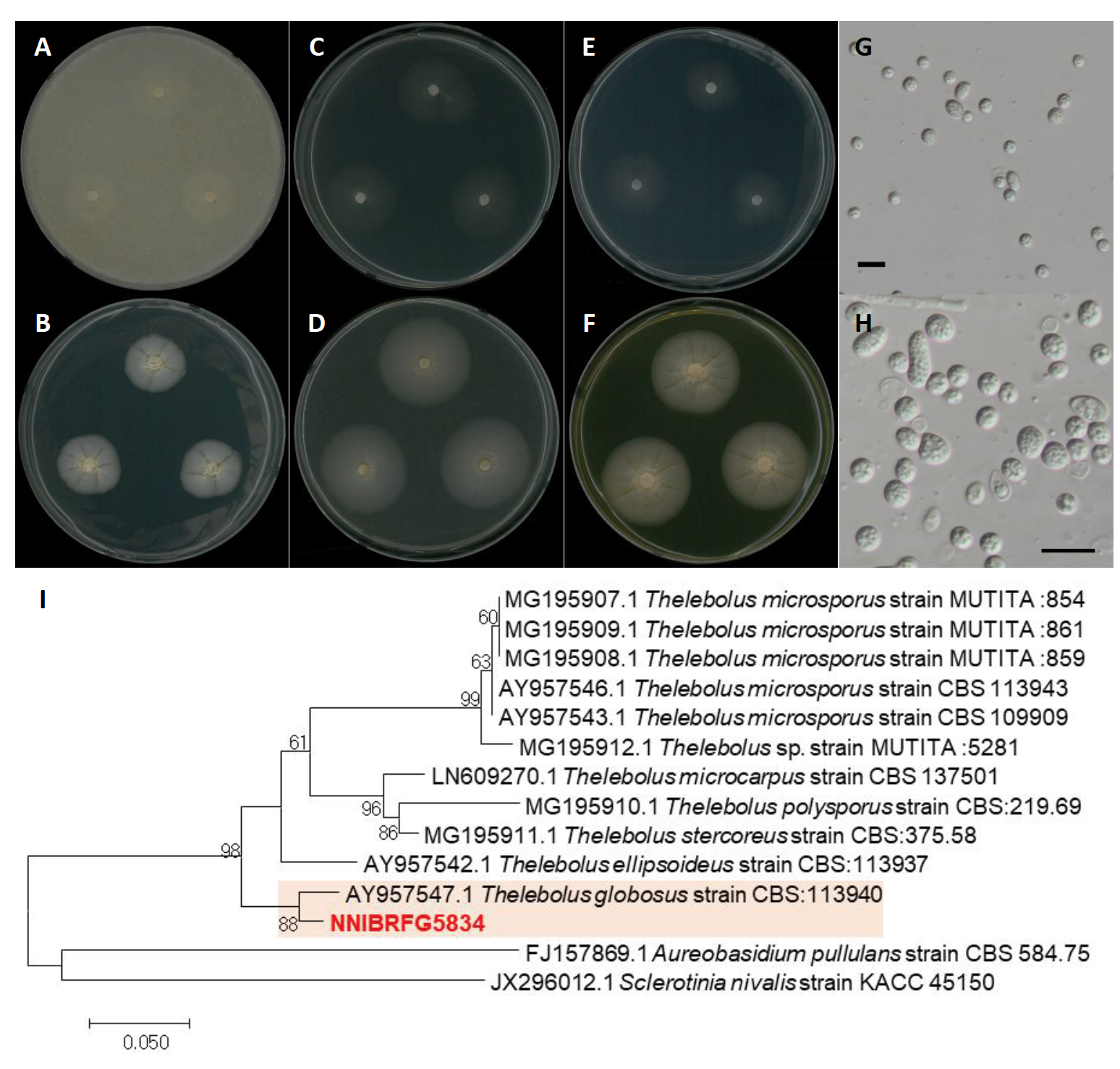
Fig. 6.Characters of Pyrenochaetopsis paucisetosa NNIBRFG27317. Mycelial growth on (A) oatmeal agar (OA), (B) potato carrot agar (PCA), (C) potato dextrose agar (PDA), (D) yeast peptone dextrose agar (YPDA), (E) malt extract agar (MEA), (F) V8-juice agar (V8A) for 7 days at 25℃. Microscopic observation showed (G-H) conidia morphology (scale bars-10 μm). Phylogenetic tree of Pyrenochaetopsis paucisetosa NNIBRFG27317 and related species, based on maximum-likelihood analysis of a combination of (I) internal transcribed spacer (ITS) rDNA and 28S rDNA sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.
Plagiostoma mejianum D.M. Walker & B.R. Lawr., Mycological Progress 13: 1061 (2014) [MB#806100] (Fig. 7)
Description: Colonies grew at a medium rate and reached a diameter of 39 mm on PDA, 29 mm on OA, 33 mm on MEA, and 29 mm on CMDA at 25℃, 7 days after inoculation. The color of the colony was pale lavender grey to white at the wavy margins with fluffy aerial mycelium on PDA; hyaline white with smooth aerial mycelium on OA; hyaline white to olive green with concentric smooth aerial mycelium on MEA; hyaline white to olive green with smooth aerial mycelium on CMDA. Conidiomata and conidia were observed on OA 21 days after inoculation and growth at 25℃. Conidia were fusiform, hyaline, one-septate, cylindrical, and measuring 12.3-20.9 µm×2.6-5.3 μm (x=17.08±1.65 µm×3.85±0.59 μm, L/W ratio=4.43, n=50).
Habitat: Plant litter in freshwater
Specimen examined: Gaegokcheon, Gaegok-li, Gapyeong-eup, Gapyeong-gun, Gyeonggi-do, 28 Mar 2019, NNIBRFG22701, Nakdonggang National Institute of Biological Resources
Note: The type material of P. mejianum, CBS 113940, was isolated from overwintered twigs of Salix babylonica [21]. In the present study, NNIBRFG22701 was isolated from plant litter in freshwater. Since the asexual stage of this species had not been previously described, this is the first report of asexual conidia in P. mejianum.
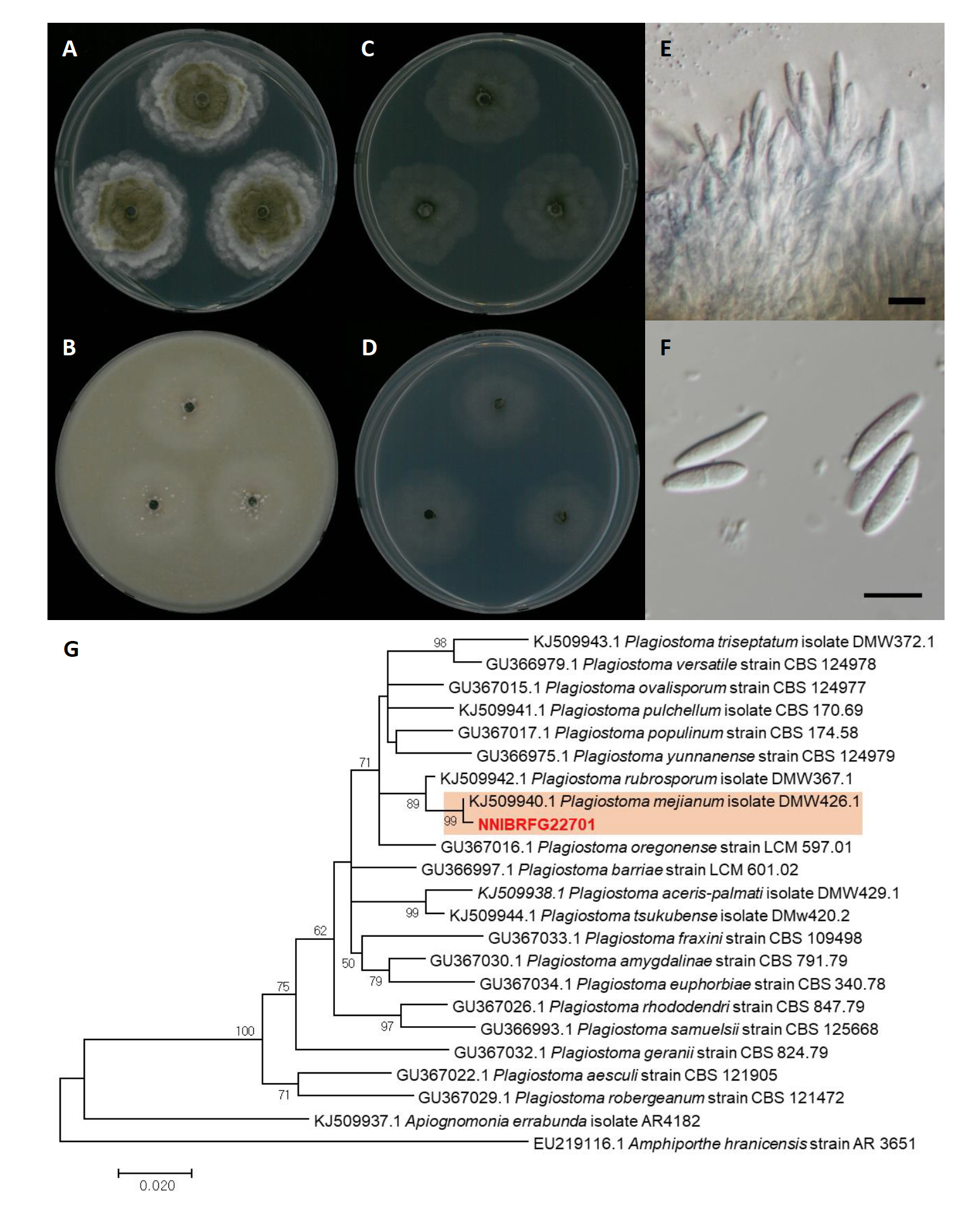
Fig. 7. Characters of Plagiostoma mejianum NNIBRFG22701. Mycelial growth on (A) potato dextrose agar (PDA), (B) oatmeal agar (OA), (C) malt extract agar (MEA )(,D) corn meal dextrose agar (CMDA ) for 7 days at 25℃. Microscopic observation showed (E) conidiophore and (F) conidia morphology (scale bars, 10 μm). Phylogenetic tree of Plagiostoma mejianum NNIBRFG22701 and related species, based on maximum-likelihood analysis of (G) the beta-tubulin sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold.
Trichoderma cremeum P. Chaverri & Samuels, Studies in Mycology 48: 63 (2003) [MB#488508] (Fig. 8)
Description: Colonies grew fast and reached a diameter of 87 mm on PDA, 87 mm on SNA, 87 mm on CMDA at 25℃, 7 days after inoculation. The color of the colony was white with wooly floccose aerial mycelium on PDA; hyaline to white with feathery aerial mycelium on SNA; hyaline with flat aerial mycelium on CMDA. Conidiation were observed 2-3 days after innoculation, and turn to green after 4-5 days. Conidia were green, smooth, obovoid to subglobose, and measuring 3.4-5.4 µm×3.1-5.0 μm (x=4.56±0.49 µm×4.06±0.56 μm, L/W ratio=1.12, n=50).
Habitat: Sediment in a stream
Specimen examined: Wamokgol, Jicheon-dong, Sangju-si, Gyeongsangbuk-do, 19 Jun 2019, NNIBRFG22286, Nakdonggang National Institute of Biological Resources
Note: Previous reported T. cremeum strains were isolated from decorticated wood [23]. In this study, NNIBRFG22286 was isolated from freshwater sediment.
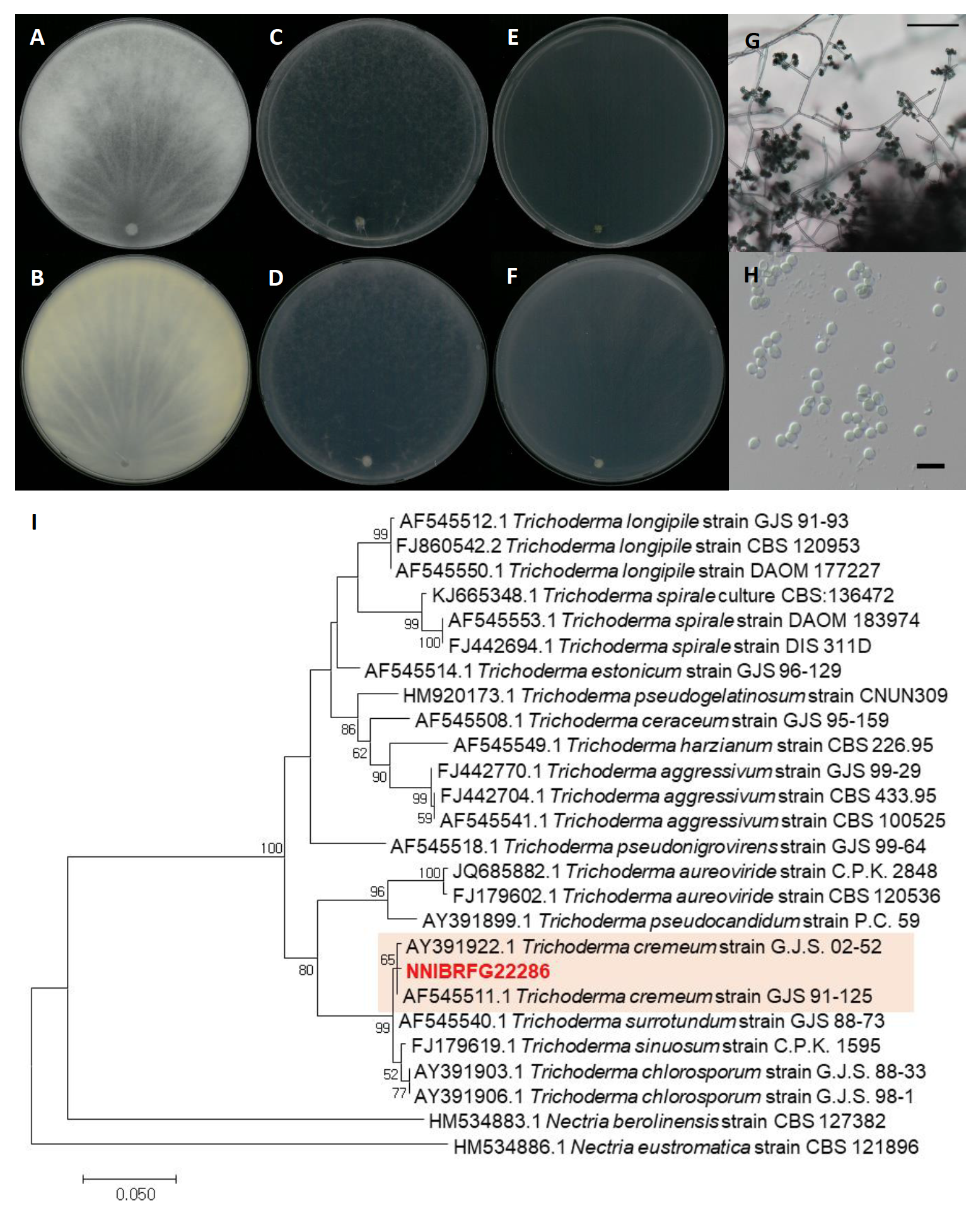
Fig. 8.Characters of Trichoderma cremeum NNIBRFG22286. Mycelial growth on potato dextrose agar (PDA; A, front; B, back), synthetic nitrogen-poor or nutrient-poor agar (SNA; C, front; D, back), corn meal dextrose agar (CMDA; E, front; F, back) for 7 days at 25℃. Microscopic observation showed (G) conidiophore and (H) conidia morphology (scale bars, G-100 μm, H-10 μm). Phylogenetic tree of Trichoderma cremeum NNIBRFG22286 and related species, based on maximum-likelihood analysis of (I) the RNA polymerase II subunit 2 sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold.
Fusarium tanahbumbuense Maryani, Sand.-Den., L. Lombard, Kema & Crous, Persoonia 43: 63 (2019) [MB#828962] (Fig. 9)
Description: Colonies grew fast and reached a diameter of 44 mm on PDA, 64 mm on OA, 60 mm on MEA, 59 mm on CMDA, and 61 mm on YPDA at 25℃, 7 days after inoculation. The color of the colony was white to creamish with wooly floccose aerial mycelium on OA; translucent white to ivory white with fluffy aerial mycelium on PDA; hyaline to yellowish white with smooth surface on MEA; white to yellowish white with dense floccose aerial mycelium on YPDA; and hyaline to yellowish white with rare aerial mycelium on CMDA. Conidiophores on aerial mycelium was abundant on PDA. Conidia were falcate, 3-5-septate, and measuring 13.6-51.7 µm×2.9-6.7 μm (x=33.35±8.09 µm×4.67±0.80 μm, L/W ratio=7.13, n=50).
Habitat: Sediment in wetland
Specimen examined: Suncheonman wetland, Suncheon-si, Jeollanam-do, Republic of Korea, 19 Oct 2019, NNIBRFG27319, Nakdonggang National Institute of Biological Resources
Note: The type material of F. tanahbumbuense, InaCC F965, was isolated from infected pseudoterm of Bumbu [24]. In the present study, NNIBRFG27319 was isolated from wetland sediment.
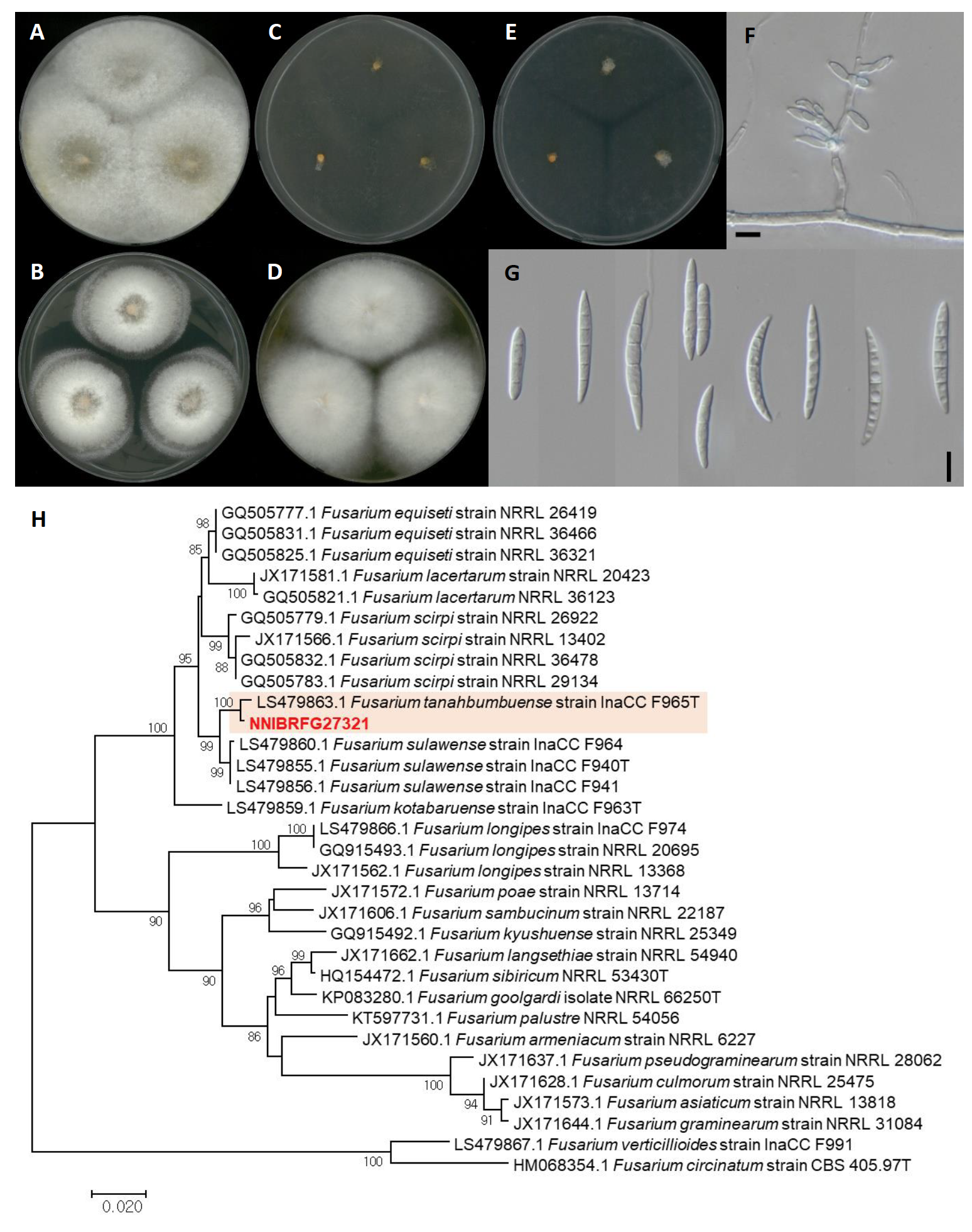
Fig. 9.Characters of Fusarium tanahbumbuense NNIBRFG27321. Mycelial growth on (A) oatmeal agar (OA), (B) potato dextrose agar (PDA), (C) malt extract agar (MEA), (D) yeast peptone dextrose agar (YPDA), (E) corn meal dextrose agar (CMDA) for 7 days at 25℃. Microscopic observation showed (F) conidiophore and (G) conidia morphology (scale bars, 10 μm). Phylogenetic tree of Fusarium tanahbumbuense NNIBRFG27321 and related species, based on maximum-likelihood analysis of (H) the RNA polymerase II subunit 2 sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.
Coniochaeta endophytica A.H. Harr. & A.E. Arnold, Plant and Fungal Systematics 64 (1): 74 (2019) [MB#830070] (Fig. 10)
Description: Colonies grew slowly and reached a diameter of 17 mm on PDA, 23 mm on MEA, and 8 mm on SNA at 25℃, 7 days after inoculation. The color of the colony was translucent white to ivory white with dense aerial mycelium on PDA; hyaline to white with smooth surface on MEA; hyaline to yellowish white with rare aerial mycelium on SNA. Conidia were aseptate, hyaline, cylindrical, ellipsoidal to fusiform, and measuring 2.6-13.1 µm×1.1-5.6 μm (x=5.30±2.82 µm×2.55±1.37 μm, L/W ratio=2.07, n=50).
Habitat: Plant litter in freshwater
Specimen examined: Wicheon, Iyeon-li, Danbuk-myeon, Uiseong-gun, Gyeongsangbuk-do, 05 Dec 2018, NNIBRFG15250, Nakdonggang National Institute of Biological Resources
Note: The type material of C. endophytica, AEA 9094, was isolated as an endophyte from healthy photosynthetic tissue of Platycladus orientalis [25]. In this study, NNIBRFG15250 was isolated from plant litter in freshwater.
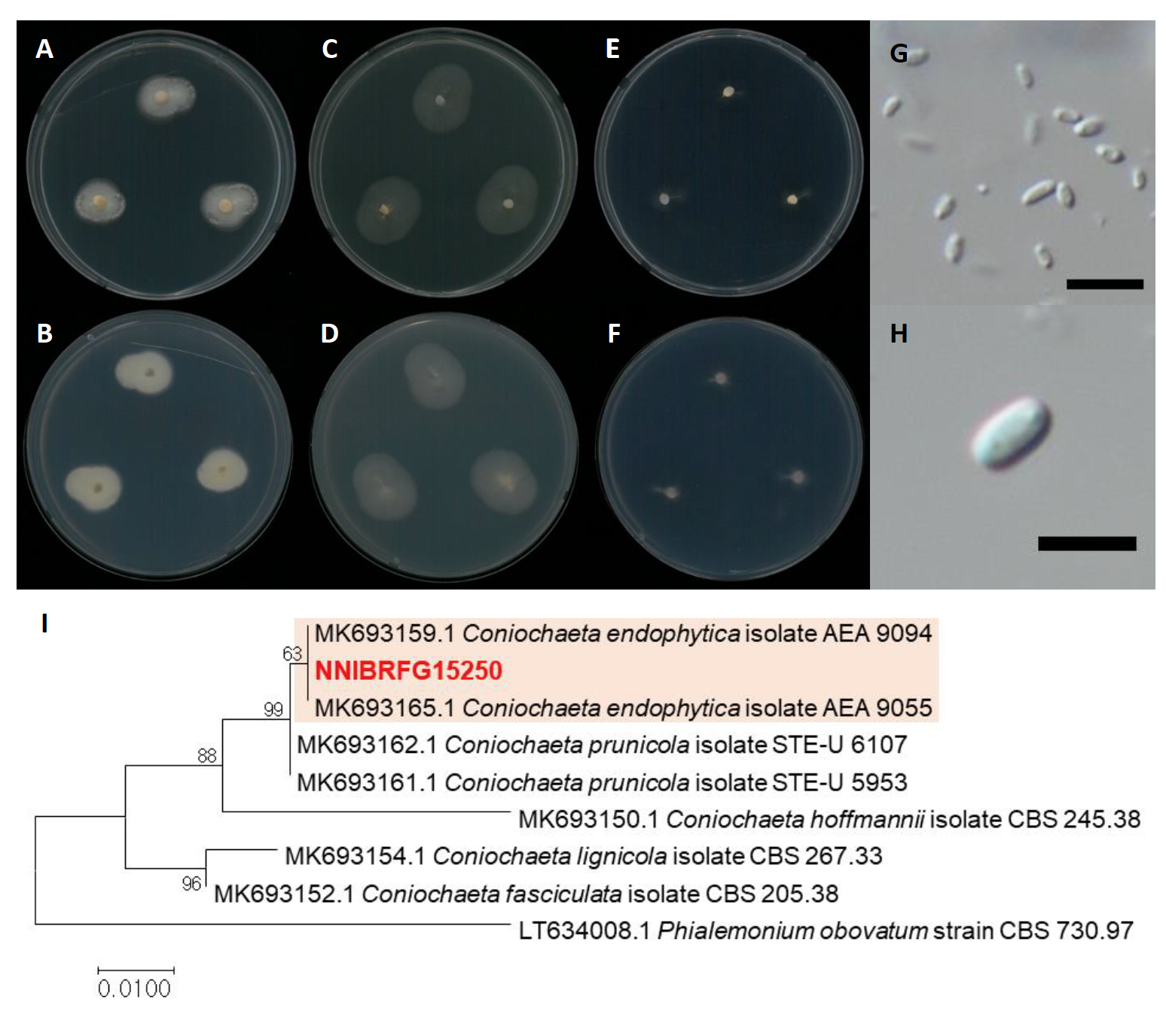
Fig. 10.Characters of Coniochaeta endophytica NNIBRFG15250. Mycelial growth on potato dextrose agar (PDA; A, front; B, back), malt extract agar (MEA; C, front; D, back), synthetic nitrogen-poor or nutrient-poor agar (SNA; E, front; F, back) for 7 days at 25℃. Microscopic observation showed (GH) conidia morphology (scale bars, G-10 μm, H-5 μm). Phylogenetic tree of Coniochaeta endophytica NNIBRFG15250 and related species, based on maximum-likelihood analysis of the translation elongation factor 1 (EF1) sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold.
Chaetomium tenue X.W. Wang, Crous & L. Lombard, Persoonia 36: 125 (2015) [MB#812986] (Fig. 11)
Description: Colonies grew fast and reached a diameter of 63 mm on PDA, 66 mm on OA, 63 mm on MEA, and 21 mm on SNA at 25℃, 7 days after inoculation. The color of the colony was hyaline yellowish olive with sparse aerial mycelium on MEA; yellow green with fluffy aerial mycelium on PDA; yellowish white to olivaceous with cottony aerial mycelium on OA; and translucent yellowish white with an uneven margin and flatted aerial mycelium on SNA. Asci were fasciculate, clavate or slightly fusiform, with eight biseriate ascospores. Ascospores were brown, elongate limoniform to broadly fusiform, with an apical germ pore, and measuring 3.2-11.3 µm×2.1-8.7 μm (x=8.32±1.70 µm×6.22±1.40 μm, L/W ratio=1.33, n=50).
Habitat: freshwater
Specimen examined: Yongjeoncheon, Juwangsan-myeon, Cheongsong-gun, Gyeongsangbuk-do, Republic of Korea, 14 Jun 2019, NNIBRFG22929, Nakdonggang National Institute of Biological Resources
Note: There is no available information on the source and geographic location of isolation of the type material of C. tenue, CBS 139.38 [27]. An asexual morph of this species was not known; consistently, in the present study the Korean strain NNIBRFG22929, isolated from an environmental sample (water), showed only a sexual morph.
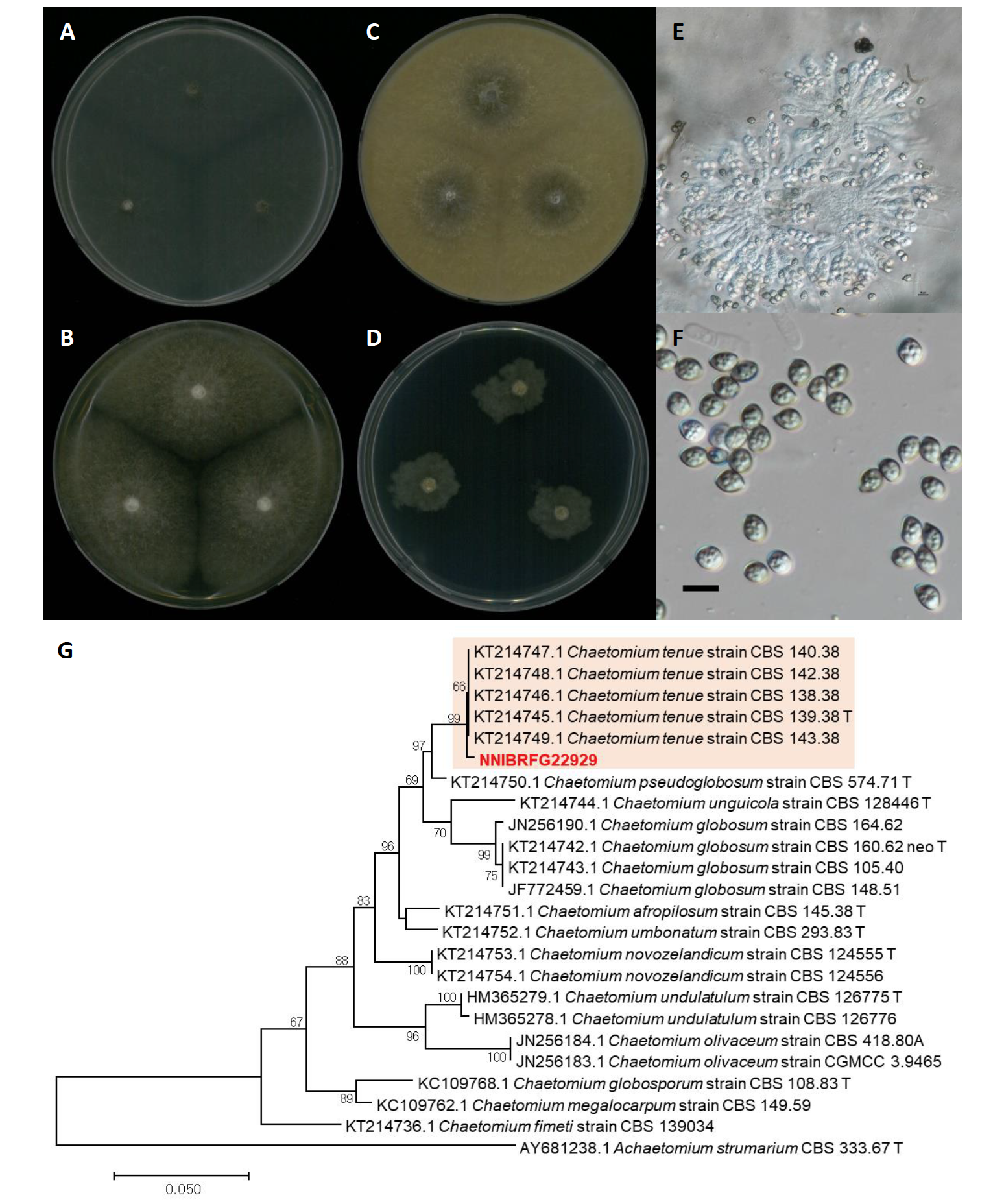
Fig. 11.Characters of Chaetomium tenue NNIBRFG22929. Mycelial growth on on (A) malt extract agar (MEA), (B) potato dextrose agar (PDA), (C) oatmeal agar (OA), (D) synthetic nitrogen-poor or nutrient-poor agar (SNA) for 7 days at 25℃. Microscopic observation showed (E) ascus and (F) ascospore morphology (scale bars, G-50 μm, F-10 μm). Phylogenetic tree of Chaetomium tenue NNIBRFG22929 and related species, based on maximum-likelihood analysis of (G) the beta-tubulin sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. New isolates from the present study are shown in bold. “T” means type material.