Abstract
This study focused on the isolation and characterization of wild yeasts in Korea. The yeasts were identified by phylogenetically analyzing the D1/D2 domains of the 26S rDNA regions. Consequently, we identified seven strains, NNIBRFG856, NNIBRFG3732, NNIBRFG3734, NNIBRFG3738, NNIBRFG3739, NNIBRFG5497, and NNIBRFG6049, which were confirmed
to be
Figures & Tables
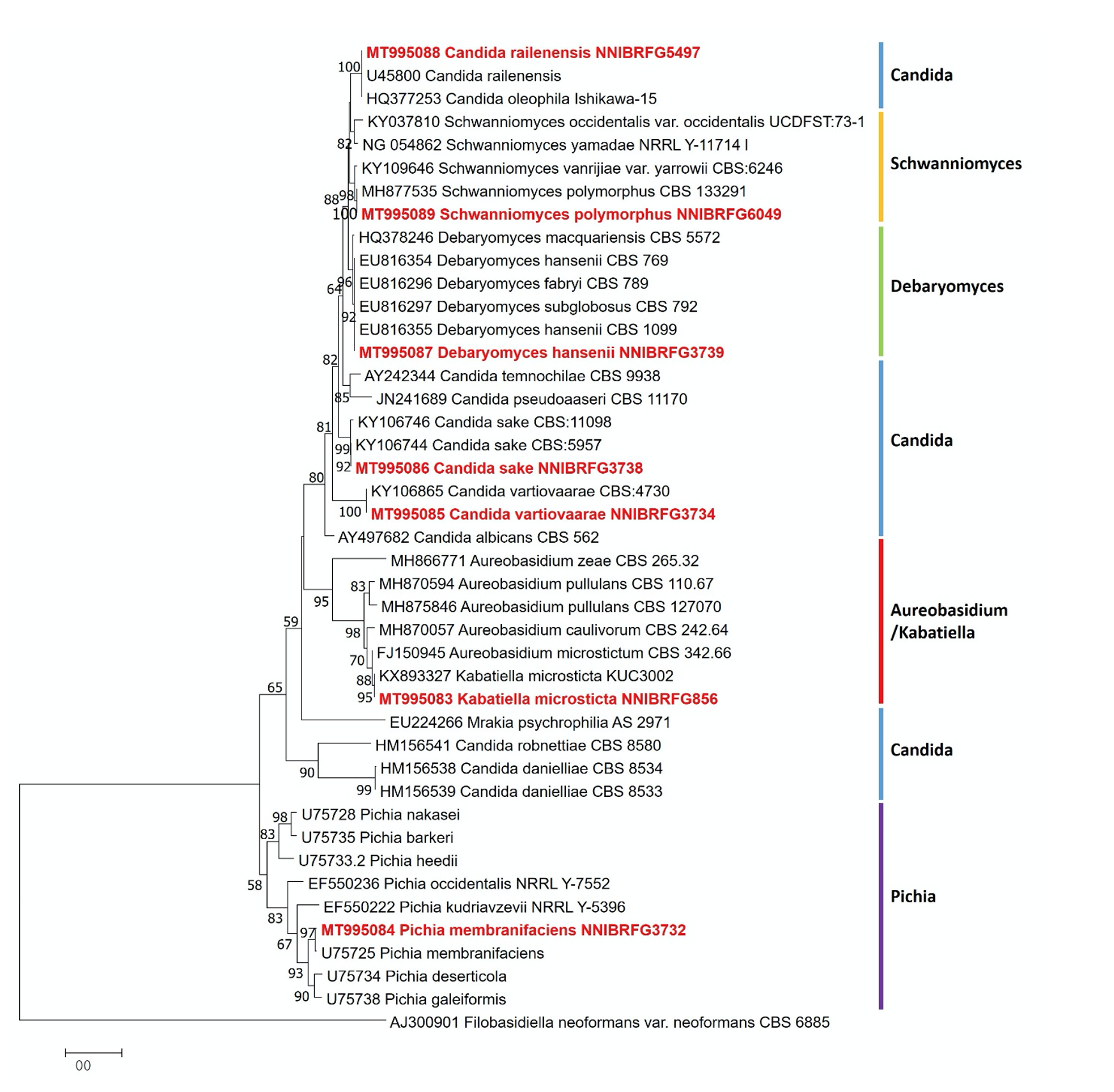
Fig. 1.Phylogenetic tree of NNIBRFG3732, NNIBRFG3734, NNIBRFG3738, NNIBRFG5497, NNIBRFG3739, NNIBRFG6049, NNIBRFG856 and related species based on a Neighbor-joining analysis of 26S rDNA sequences. The sequence of var. was used as an outgroup. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. The bar indicates the number of substitutions per position. The new isolates from the present study are shown in bold and red.


