INTRODUCTION
The class Dothideomycetes is the largest and most diverse Ascomycota containing 11 orders, 90 families, 1,300 genera, and over 19,000 known species [1,2]. The order Dothideales, that belongs to the class Dothideomycetes, was introduced by Lindau (1897) to provide a single-family of Dothideaceae. Since then, the family Dothideaceae had numerous changes in its genera concept and inclusion from 1896 to early 2000, including ascomata developing nonostiolate loculi in stromata, opening by apical fissure or dehiscence, displaying eight-or many-spored asci at the base of the locules with one or many-septate, a different hyaline or brown appearance, and presenting guttulate ascospores [3]. Dothidea insculpta is a fungal species in the family Dothideaceae. The family Dothideaceae is composed of fungi that are biotrophic, necrotrophic, or saprobic. These fungi infect twigs and other plant parts, as well as sometimes be found on leaves as a plant pathogen that causes serious crop loss [2]. Metarhizium rileyi (Clavicipitaceae, Hypocreales, Ascomycota) under the class Sordariomycetes is widespread, and commonly known as an important entomopathogenic fungus [4]. Yeast-like hyphal bodies and a true filamentous growth phase are distinct characteristics of M. rileyi [5]. M. rileyi was described as Botrytis rileyi Farlow in 1883 originally, and later as Spicaria rileyi Farlow. In 1974, this fungus was re-described and transferred it to the genus Nomuraea Maublanc by Kish, Samson, and Allen [6]. However, it was proposed as a new combination by Kepler, Humber, Bischoff, and Rehner in 2014, moving it to the genus Metarhizium. It is also known as Nomuraea rileyi Farlow [7], as it is a key mortality factor for caterpillar populations in various crops under specific environmental conditions, including subtropical and tropical soybeans [8]. This study aimed to identify fungi isolated from soil and cherry tree (Prunus serrulata) bark. We present the morphological and molecular characteristics of two unreported fungal species namely, Dothidea insculpta and Metarhizium rileyi in Korea.
MATERIALS AND METHODS
Sample collection and fungal isolation
The soil sample from hillside root-soil and cherry tree (Prunus serrulata) bark were collected from Gyeongsangbuk-do (36°11'45.9"N, 128°34'10.8"E; 35°38'36.6"N, 128°37'31.2"E) in Korea. Bark samples were collected from the cherry tree, placed in sterile plastic bags, and taken to the laboratory for fungal isolation. A small piece of bark was cut and put onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated at 25℃. Soil samples were collected from between the surface and 20 cm below the surface, stored in a plastic bag, brought to the laboratory, and stored at 4℃ until analyzed. Then, the soil serial dilution was performed and roughly, 1 g of soil was weighed and suspended in 10 mL of sterile distilled water to make serial dilutions (10-1 to 10-5). Each soil serial dilution was vortexed, and 0.1 mL of each dilution was spread on PDA plates. Well-developed individual colonies were isolated and cultured again on fresh PDA plates and incubated at 25℃ until mycelium development. The pure cultures were preserved in 20% glycerol at -80℃ to produce a stock for future studies.
Morphological characterization
The morphological characteristics of the isolated strains were studied by growing on PDA, and malt extract agar (MEA; Difco, Detroit, MI, USA) media. The KNU-SOT5 strain was studied after 5 days at 20℃ [3]. The KNU-Gunwi 2B strain was studied after 20 days at 28℃ [4]. The cultural characteristics, colony color, texture, growth, shape, and size were studied and mycological characteristics were observed using a light microscope (BX-50; Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Fungal strains were grown on PDA plates and scraped mycelia was used for the genomic DNA extraction. Total genomic DNA was extracted using a HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s instructions. The polymerase chain reaction (PCR) was carried out using three partial gene portions in this study. NS1 and NS4 primers were used to amplify a region spanning the small subunit (SSU) rDNA [9]. LROR and LR5 primer pairs were used to amplify a segment of the large subunit (LSU) rDNA [10] and internal transcribed spacers (ITS) were amplified by using the primer pairs ITS1F and ITS4 [9,11]. The amplified PCR products were purified using EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Solgent Co., Ltd. (Daejeon, Korea). The PCR purification products were deposited in the National Center for Biotechnology Information (NCBI) GenBank database.
Phylogenetic analysis
To make the phylogenetic tree, we analyzed the LSU, SSU, and ITS sequences for the strain KNU-SOT5, and ITS sequences for the strain KNU-Gunwi 2B and also retrieved the allied sequences from the NCBI GenBank database (Table 1). The LSU, SSU, and ITS datasets were first analyzed separately and then the individual datasets were concatenated into a combined dataset. The phylogenetic tree was constructed based on the maximum likelihood algorithm using the Kimura model and the MEGA 7.0 software to bootstrap for 1,000 replications [12,13].
RESULTS AND DISCUSSION
Dothidea insculpta Wallr., Flora Cryptogamica Germaniae 2: 864 (1833) [MB#173197] (Fig. 1)
Specimen examined: Cheongdo-gun, Gyeongbuk province (35°38'36.6"N, 128°37'31.2"E), isolated from cherry tree (Prunus serrulata) bark. The stock culture (NREFFGC000000241) was deposited in the National Institute of Biological Resources (NIBR) as a metabolically inactive culture.
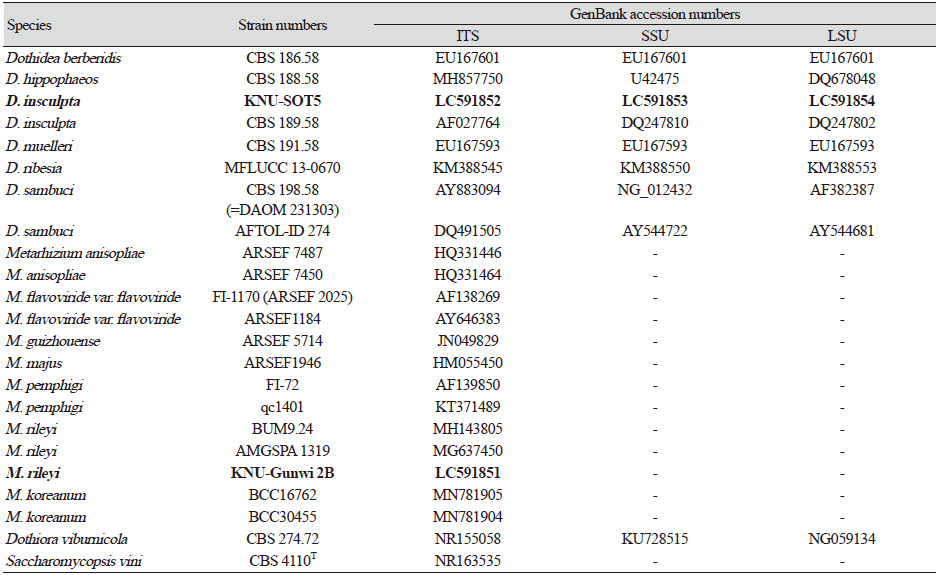
Fig. 1. Morphological characteristics of Dothidea insculpta KNU-SOT5. A, colonies on potato dextrose agar (PDA) after 5 days; B, large colonies on PDA after 2 wks; C, feathery mycelium and blackish conidiomata-like structures observed under a stereomicroscope; D, E, conidiophores; F, melanized hyphae developing into chlamydospores; G, conidia. Scale bars: C=50 μm; F=20 μm, D, E=10 μm, G=5 μm.
Morphological characteristics of KNU-SOT5
The colonies were rhizoid, flat, rough-surfaced, and had sparse to moderate aerial mycelium with feathery rhizoid margins. Colony diameter reached 24 mm on PDA and 11 mm on MEA media after 5 days at 20℃. On PDA, the colony upper surface was white around the margin, which turned to greenish-black as approaching the center; the back color was black (Fig. 1A). The rhizoid form colony was more clearly observed after 2 weeks of incubation (Fig. 1B). Hyphae were sub-hyaline to brown, branched, thin-walled, septate. The strain also produced feathery mycelium and blackish conidiomata-like structures on the agar surface (Fig. 1C). Conidiophores were observed, which arise directly from the mycelia and short side-branches with top swollen, smooth-walled, hyaline (Fig. 1D and 1E). Chlamydospores were dark brown, smooth to lightly rough-walled, and globose to ellipsoidal (Fig. 1F). Conidia were aseptate, guttulate, variable in shape, but mostly fusiform, and 5.3-17.6×4.2-7.0 μm in size (Fig. 1G). The culture and morphological characteristics of the isolated strain KNU-SOT5 suggested that the fungus was most closely related to Dothidea insculpta.
Molecular phylogeny of the KNU-SOT5
Sequences containing 582, 991, and 829 bp were obtained which corresponded to the ITS, SSU, and LSU, accordingly. According to result of Basic Local Alignment Search Tool (BLAST) in NCBI, the obtained sequence showed 99.4, 99.9, and 99.9% similarities with the strain of Dothidea insculpta CBS 189.58 from the sequences of ITS, SSU, and LSU, accordingly. A phylogenetic tree was constructed based on the combined sequences of ITS, SSU, and LSU regions by using a maximum likelihood method. The strain KNU-SOT5 was clustered together with D. insculpta CBS 189.58 showing 87% bootstrap values (Fig. 2).
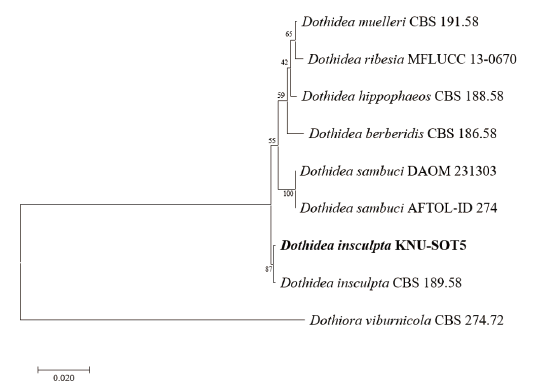
Fig. 2. Maximum likelihood phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) region, large subunit rRNA (LSU) and small subunit rRNA (SSU) gene. Dothiora viburnicola CBS 274.72 was used as an out-group. The strain isolated in this study is in bold, and the bootstrap values are based on 1,000. Bar, 0.02 substitutions per nucleotide position.
In the previous study, D. insculpta was isolated from a dead branch of Clematis vitalba tree which is under the family Ranunculaceae from Italy [3]. Dothidea species are widely distributed around the world, especially in tropical areas [14]. And also, another species belongs to the genus Dothidea, namely D. cladonema (=Clypeococcum cladonema) found on volcanic rocks from France [15], D. eucalypti on leaves of Eucalyptus dalrympleana (Myrtaceae) in Australia [16], D. sambuci on Sambucus nigra (Adoxaceae) in Austria [3]. Though, there are so many Dothidea species isolated from different countries, but still now, there is no study in Korea. However, during fungal diversity surveys, there were two fungal strains, namely Paecilomyces variotii and Talaromyces amestolkiae were isolated from samples of rat dung and fig tree leaf collected at a garden located in Gwangju in Korea [17]. A disease also called "Brown rot" was discovered to be present on cherry fruits (Prunus avium L.) in the city of Hwaseong, Korea [18]. In this study, the strain KNU-SOT5 was collected from Gyeongsangbuk-do associated with the cherry tree (Prunus serrulata) bark in Korea. Thus, further research is being required for this species in future, not only a better understanding of the fungal disease of cherry tree but also the classification and ecology that are mostly important in Korean environmental conditions. To our knowledge, this is the first report of Dothidea insculpta in Korea.
Metarhizium rileyi (Farl.) Kepler, S.A. Rehner & Humber, Mycologia 106 (4): 824 (2014) [MB#807862] (Fig. 3)
Specimen examined: Gunwi-gun, Gyeongbuk province (36°11'45.9"N, 128°34'10.8"E), isolated from hillside root-soil. The stock culture (NREFFGC000000235) was deposited in the National Institute of Biological Resources (NIBR) as a metabolically inactive culture.
Morphological characteristics of KNU-Gunwi 2B
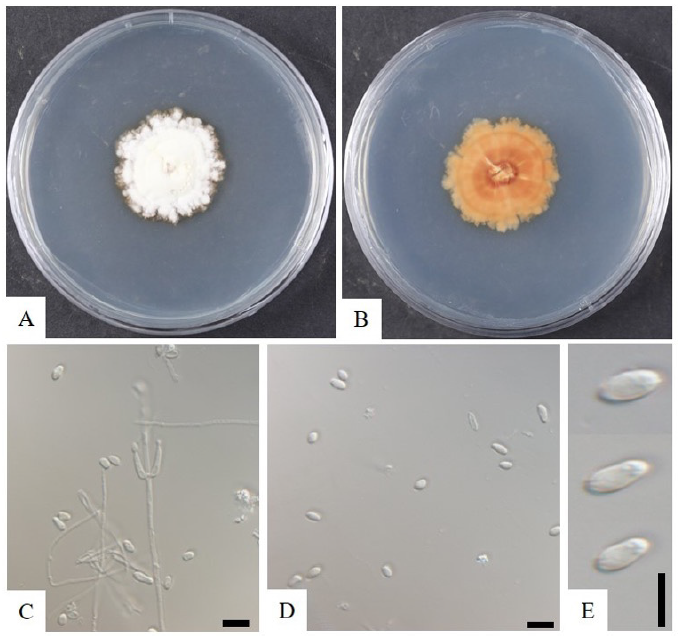
The strain KNU-Gunwi 2B was cultured on PDA media for 20 days at 28℃ to study the cultural and morphological characteristics (Fig. 3A and 3B). The colony diameter reached 24 mm on PDA plates with the slow growth rate, and the color was white with irregular borders that turned to pale-green; the back color was pale yellow. The hyphae were up to 2.4 µm in width, hyaline, septate, and smooth. The conidiophores were hyaline, erect, septate, originating from hyphae with two phialides (Fig. 3C). Phialides were smooth-walled, cylindrical with semi-papillate apices, and 10.0-12.0×2.0-3.0 μm in size. The conidia were broadly ellipsoid, sometimes cylindrical, and with the diameter of 4.4-8.0×2.3-4.0 μm (Fig. 3D and 3E).
Molecular phylogeny of the KNU-Gunwi 2B
From the sequence analysis, a 632 bp sequence was obtained from ITS regions. The ITS sequence had 99.8% similarities with the different strains of Metarhizium rileyi (MH143805, AB268359, and MG637450). The ITS regions sequences were used to construct a phylogenetic tree by using the Maximum Likelihood method with the retrieved allied species sequences from the NCBI database. The phylogenetic tree revealed that the strain KNU-Gunwi 2B was clustered with different strains of M. rileyi BUM 9.24 and M. rileyi AMGSPA 1919 with 100% bootstrap values (Fig. 4).
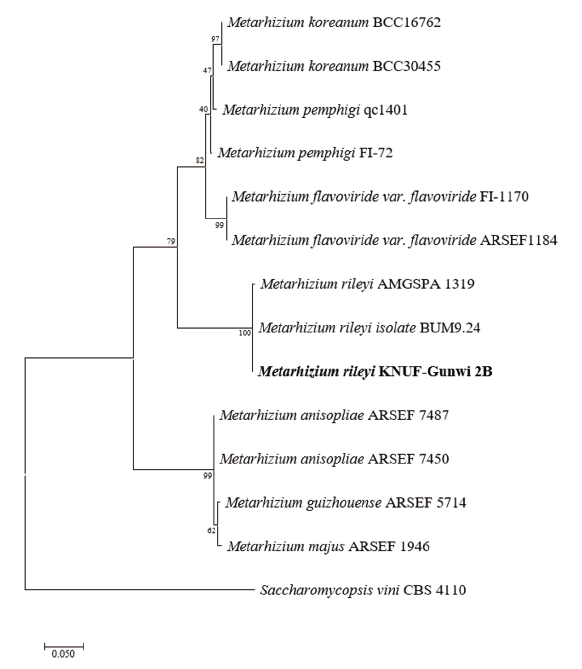
Fig. 4. Maximum likelihood phylogenetic tree analysis based on Metarhizium rileyi KNU-Gunwi 2B internal transcribed spacer rDNA sequences. Saccharomycopsis vini CBS 4110 was used as an outgroup. The strain isolated in this study is in bold, and the bootstrap values are based on 1000 replications. Bar, 0.05 substitutions per nucleotide position.
There are many new species were added to Metarhizium in the last five years [19-24]. Most well-known Metarhizium species were identified as entomopathogens. Some were also reported as endophytes (soil and rhizosphere inhabitants) [25-27], resulting in increased plant growth and providing increased pest and disease tolerance [28,29]. Metarhizium were also reported in China, Japan, and Thailand mainly from insects, soils, and plant roots [28,30]. Moreover, there are some species belonging to the genus Metarhizium isolated from diversified hosts and habitats from different countries such as M. anisopliae (insects from Orthoptera, Coleoptera-Brazil, India, Eritrea, Eastern Africa), M. flavoviride var. flavoviride (insects from Coleoptera, Agricultural Soil-France, Germany, Netherlands), M. flavoviride var. minus (insects from Homoptera-Philippines, Solomon Islands), M. frigidum (insects from Coleoptera, Soil, Termite mound-Australia) [31]. However, the fungal strains were also isolated from environmental soil that was found in various agricultural areas, namely Penicillium raphiae [32], Metarhizium guizhouense and Mortierella oligospora in Korea [33]. In the present study, the strain KNU-Gunwi 2B was isolated from hillside root-soil in Gyeongsangbuk-do also and identified as Materhizium rileyi.
In conclusion, further research is being required for these two species in the future not only to have a better understanding of soil and cherry tree fungal diseases but also about these species classification and ecology that are mostly important in agricultural production in Korean environmental conditions. To our knowledge these two species, namely Dothidea insculpta KNU-SOT5 and Metarhizium rileyi KNU-Gunwi 2B, are the first reports in Korea.