Introduction
Endophytic fungi colonize the tissues of most plants without causing disease symptoms [1,2]. They typically provide benefits-such as tolerance to environmental stresses, pathogen resistance, and production of secondary metabolites-to their hosts. Endophytic fungi are found in all parts of host plants, including the leaves, stems, and roots. Root-endophytes differ substantially in function from shoot-endophytes and are similar to mycorrhizal fungi. In terms of their function, however, mycorrhizal fungi form extra-root hyphae to transport nutrients to the host plants [3], whereas root endophytes, instead use enzymes to convert organic nitrogen and phosphorus into inorganic matter, which is advantageous for obtaining nutrients around plant roots [4,5]. Root endophytic fungi, including dark septate endophytes and Trichoderma, are diverse and different from the fungi colonizing aboveground plant tissues. This study investigated endophytic fungal diversity in the roots of Quercus spp. in Korea. As a representative tree genus, Quercus spp. account for approximately 20% of the forest area in Korea. We isolated four species of endophytic fungi from the roots of three Quercus species and identified them based on their morphological and molecular characteristics. To the best of our knowledge, this is the first report on these four fungal species in Korea.
Materials and Methods
The roots of Quercus spp. were collected from three sites, Jecheon, Danyang, and Gyeongju. The samples were transported to the laboratory in polyethylene bags within 24 h of collection. Healthy roots without disease symptoms were selected and washed with tap water before being surface sterilized with 1% NaClO solution for 1 min and 70% EtOH for 2 min [6]. The surface-sterilized roots were then cut into 1.5-2.0 cm sections, placed in Petri-dishes containing potato dextrose agar (PDA) medium, and incubated at 25°C [7]. Hyphae growing from root fragments were transferred to new Petri dishes containing PDA and incubated at 25°C. Fungal isolates were deposited at the Culture Collection of the National Institute of Biological Resources (NIBR), Incheon, Korea.
The growth characteristics of fungal colonies were recorded after incubation on PDA at 25°C for 7 days. Conidiophores and conidia were examined under a light microscope (AXIO Imager A1; Carl Zeiss, Oberkochen, Germany). Genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA). Internal transcribed spacer (ITS) and large subunit (LSU) regions of rDNA were amplified using the primers ITS1F/ITS4 [8] and LROR/LR16 [9], respectively. Amplicons were sequenced by SolGent Co. (Daejeon, Korea) and then sequences were deposited in the NCBI GenBank. Neighbor-joining phylogenetic analysis was conducted using the combined ITS and LSU sequences in MEGA10, based on the Kimura-2 parameter distance model [10]
Results and Discussion
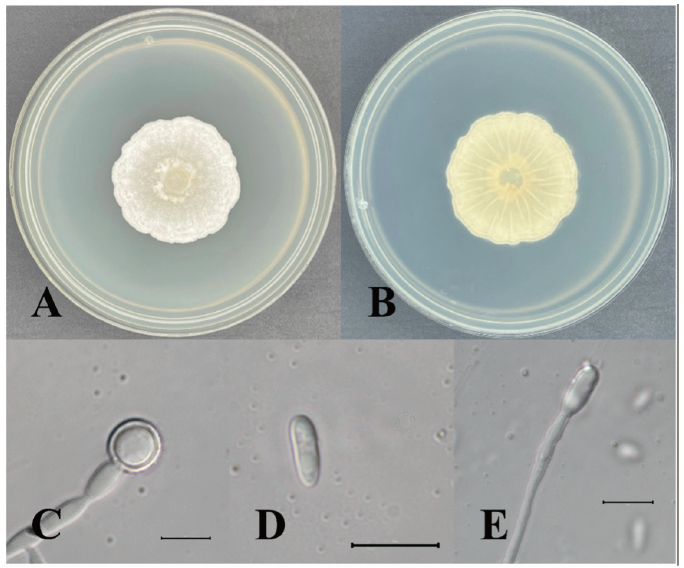
Acremonium crotocinigenum (Schol-Schwarz) W. Gams, Cephalosporium-artige Schimmelpilze:112 (1971) [MB#308136] (Fig. 1, Fig. 2, Table 1)
Morphological characteristics of KNUE20T014: Colony diameter was 37–39 mm after 7 days at 25°C on PDA. The front side was ivory in color, and the back side was pale yellow. Exudates were absent, texture was downy, and margins were circular. Conidia were (5.11-) 6.55 (-8.57) × (2.61-) 3.17 (-4.0) µm in size, with numerous hyaline features. Chlamydospores were round or oval, up to 12 µm in diameter
Specimen examined: Danyang-gun, Chungcheongbuk-do, Republic of Korea, 37°57'N, 124°38'E, 4. 23. 2020, isolated from the roots of Quercus dentata, NIBR No. NIBRFGC000508406, GenBank No. MZ895482.
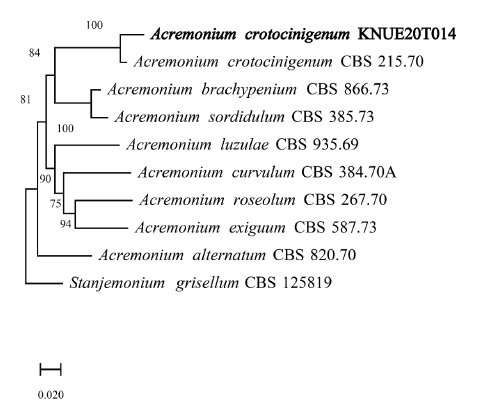
Note: A. crotocinigenum was first described by Schol-Schwarz as Cephalosporium crotocinigenum and named after its production of crotocin mycotoxin [11]. It is distinguished from other Acremonium species by the presence of long muti-septate conidiophores and chlamydospore [11]. BLAST analysis revealed that the ITS sequence of KNUE20T014 had 99% similarity with the A. crotocinigenum strain Dzf16 (EU543259.1) and the LSU region had 99% similarity with strain CBS 408.70A (MH871528.1), Neighbor joining phylogenetic analysis of the combined ITS and LSU sequences revealed that KNUE20T014 was most closely related to the A. crobociningenum strain CBS 215.70.
Table 1. Morphological characteristics of Acremonium crotocinigenum KNUE20T014 isolated in oak trees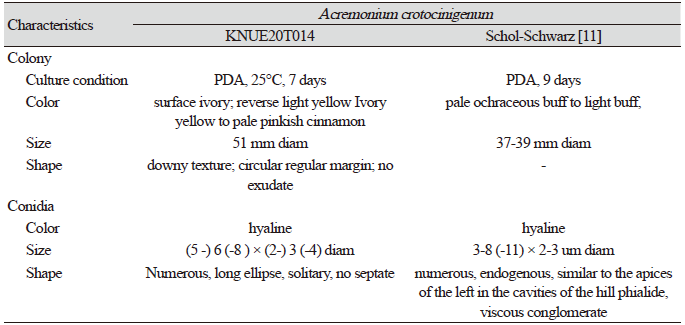
|
|
PDA, potato dextrose agar |
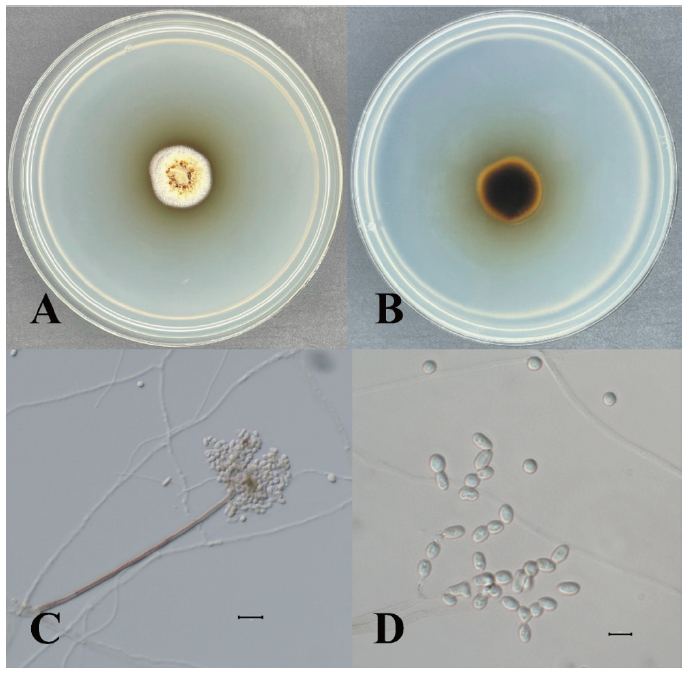
Oidiodendron citrinum G.L. Barron, Canadian Journal of Botany 40: 597 (1962) [MB#335317] (Fig. 3, Fig. 4, Table 2)
Morphological characteristics of KNUE20T046: The colony was 19–21 mm in diameter and characterized by a rounded margin. The front side was grayish and yellow-green, while the back side was dark brown with a black center and margin. The colony exhibited furrows, with no exudate. Conidia were spherical and hyaline, with a diameter of (5.79-) 7.83 (-8.80) × (4.04-) 4.97 (-5.34) µm
Specimen examined: Jecheon-si, Chungcheongbuk-do, Republic of Korea, 37°4'N, 128°12'E, 4.23. 2020, isolated from the roots of Quercus variabilis, NIBR No. NIBRFG0000509557, GenBank No. MZ895457.
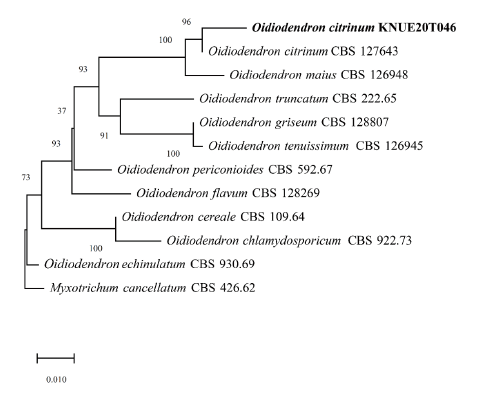
Note: Species in the genus Oidiodendron are commonly found in soils and decaying plant materials worldwide [12]. O. citrinum was first isolated by Barron in 1962 from peat soils in Ontario, Canada [12]. BLAST analysis showed that the ITS sequence was 99.28% similar between strain KNUE20T046 and strain CBS 127643 (MH864651.1), whereas the LSU of KNUE20T046 and strain CBS 129229 (MH876692.1) exhibited 100.00% similarity. Neighbor-joining analysis of the combined ITS and LSU sequences revealed that KNUE20T014 was most closely related to the O. citrinum strain CBS 127643, with a 94% bootstrap value. Rice and Currah [13] classified O. citrinum as O. maius var citrinum, based on molecular studies, suggesting that O. citrinum was not significantly different from O. maius at the species level. The latter is known to have a mycorrhizal relationship with Ericaceae. The present study confirmed that the two species were closely related, however, by comparing the morphologies of O. maius and O. citrinum, we identified strain KNUE20T046 as O. citrinum based on the yellow mycelium and rugose conidia.
Table 2. Morphological characteristics of Mammaria echinobotryoides KNUE20T102 isolated in oak
trees and previously reported isolate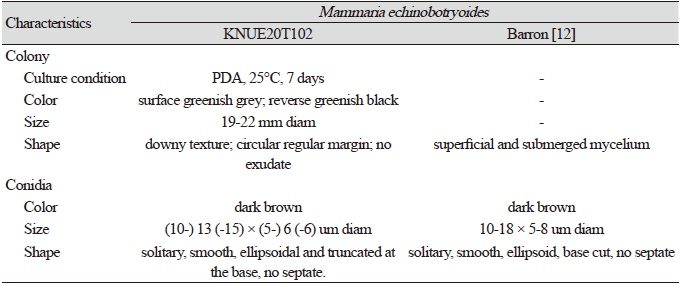
|
|
PDA, potato dextrose agar |
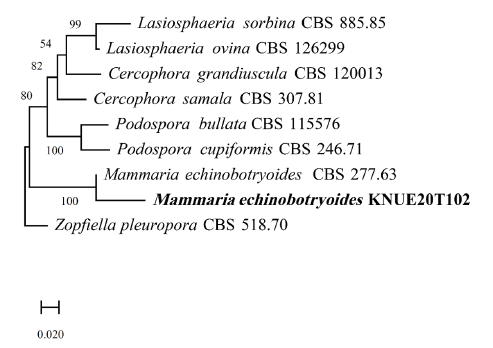
Mammaria echinobotryoides Ces., Klotzschii herbarium vivum mycologicum sistens fungorum per totam Germaniam crescentium collectionem perfectam. Cent. 19: no. 1859 (1854) [MB#179938] (Fig. 5, Fig. 6, Table 3)
Morphological characteristics of KNUE20T102: The colony was 19–22 mm in diameter, and colored greenish-light gray with dark, greenish margins. The back side was greenish-black with a greenish-gray margin. The colony had furrows; a downy texture; a rounded, regularly shaped margin; and no exudate. Conidia were smooth, oval, dark brown, lacked septa, and (10.2-) 13.5 (-15.4) × (5.0-) 6.0 (-6.8) µm in diameter.
Specimen examined: Gyeongju-si, Gyeongsangbuk-do, Republic of Korea, 35°41'N, 129°21'E, 6. 26. 2020, isolated from the roots of Quercus aliena, NIBR No. NIBRFGC000508495, GenBank No. MZ895377.
Note: Mammaria echinobotryoides was first reported by Cesati in 1854 [14]. Cesati distinguished Mammaria from two genera Echinobotryum and Wardomyces, with similar morphological characteristics, based on the development of unique conidia in Mammaria. The conidiophore of Mammaria spp. showed sympodial growth, which is consistent with the conidiophores from this study, however, those of the other two genera showed lateral or basipetal branching patterns. BLAST analysis of the ITS and LSU regions revealed 99.27% similarity with MH865136.1 and 99.51% with MH871141.1. Neighbor joining analysis of the combined ITS and LSU sequences revealed that isolate KNUE20T102 was most closely related to the M. echinobotryoides strain CBS 277.63.
Table 3. Morphological characteristics of Oidiodendron citrinum KNUE20T046 isolated in oak trees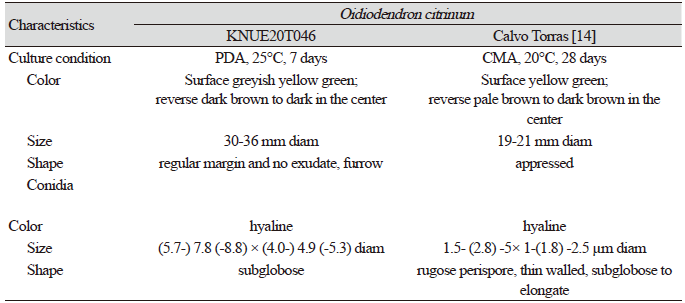
|
|
PDA, potato dextrose agar; CMA, corn meal agar |
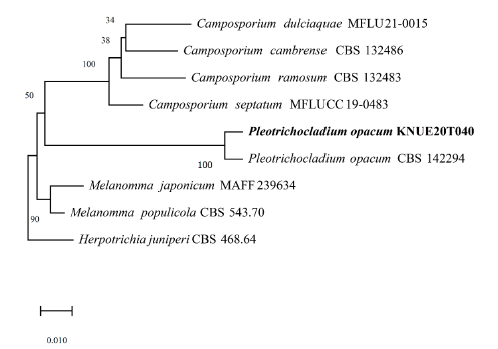
Pleotrichocladium opacum (Corda) Hern.-Restr., R.F. Castañeda & Gené, Studies in Mycology 86: 75 (2017) [MB#820278] (Fig. 7, Fig. 8, Table 4)
Morphological characteristics of KNUE20T040: The colony was 22–24 mm in diameter. The front side was greenish-brown in color, and the margin was light ivory. The back side was greenish-brown and light ivory. There were more ivory-colored areas on the margin than on the front side. A brown exudate was observed. The mycelium in the central area extended radially and formed a circular margin. The conidia were dark brown and extended from the hyphae in short branches or without branches, and (7.83-) 8.80 (-5.79) × (4.97-) 5.34 (-4.04) µm in diameter. The conidia formed club-shaped monospore with (2-) 4 (-5) septate.
Specimen examined: Jecheon-si, Chungcheongbuk-do, Republic of Korea, 37°4'N,128°12'E, 4. 23.2020, isolated from the roots of Quercus mongolica, NIBR No. NIBRFGC000508496, GenBank No. MZ895459.
Note: Pleotrichocladium opacum was first reported as Sporidesmium opacum on dead wood by Corda in 1837 and Hern established the species in the genus Pleotrichocladium via molecular phylogenetic analysis [15]. Pleotrichocladium opacum was morphologically distinguished from the closely related T. asperum, based on the color of conidia and its warty surface. BLAST analysis of the ITS and LSU regions revealed 99.84% similarity with NR_155696.1 and 99.84% similarity with KY853526.1. Neighbor-joining phylogenetic analysis of the combined ITS and LSU sequences revealed that isolate KNUE20T040 was part of the same lineage as the P. opacum CBS 142294.
Table 4. Morphological characteristics of Pleotrichocladium opacum KNUE20T040 isolated in oak trees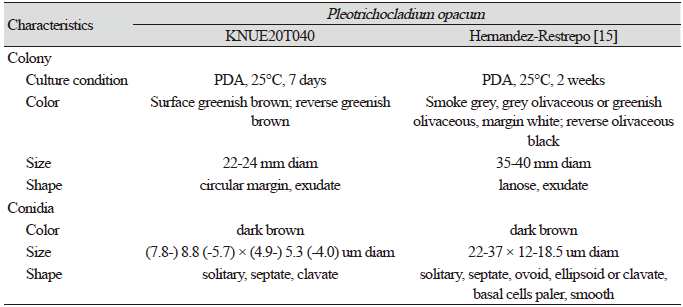
|
|
PDA, potato dextrose agar |
In this study, endophytic fungi were isolated from the roots of Quercus spp. The isolated strains were identified based on the morphological characteristics of their colonies and conidia, followed by DNA sequence analysis of the ITS and LSU regions. In this study, four species of endophytic fungi were identified; O. citrinum, A. crotocinigenum, M. echinobotryoides, and P. opacum. To the best of our knowledge, this is the first record of these four species in Korea. This study thus contributes to a greater understanding of root endophyte diversity in Korea.