Yeast is adapted to various environments and is distributed in the air, soil, and tissues of various plants and animals, including humans. In general, yeast is predominantly single-celled, and most yeasts are Ascomycetes. These yeasts are spread all over the world and are economically very important organisms [1]. Among them, the genus Galactomyces Redhead & Malloch belongs to the family Dipodascaceae and is a telemorph of the genus Geotrichum. A total of five species have been identified within the genus Galactomyces and a total of four species have been identified in its anamorph genus Geotrichum [2]. Until now, only two species have been reported in Korea: Galactomyces citri-aurantii E.E. Butler in 1958 and Geotrichum candidum Link in 2003 [3,4]. In Korea, the American crayfish (Procambarus clarkii Girard) is designated as an introduced species and an invasive species, as it has caused a massive disturbance of the Korean natural ecosystem. It also competes with the crayfish (Cambaroides similis Koelbel) distributed naturally in Korea [5]. There is no record of when or how the American crayfish was officially introduced to Korea, but it is presumed that the species was introduced as a pet and was then illegally released into the Korean natural ecosystem by humans [5].
In the National Institute of Ecology (NIE), some experts in crustaceans are investigating the competition between American crayfish and crayfish, their effect on the Korean ecosystem, and control methods. In line with those investigations, we conducted a pilot study on the biodiversity of the fungi inhabiting the American crayfish and discovered Galactomyces pseudocandidus de Hoog & M.T. Sm. in its digestive tract. Therefore, based on the morphological and molecular characteristics, we confirmed that it is an unrecorded fungus in Korea, and we report this species here.
The American crayfish used in the study were collected from Mosan reservoir in Hampyeong-gun in 2021. The collected species were dissected, and the digestive tract was extracted for fungal isolation. The extracted digestive tract was prepared by washing three times with sterile water and then placed in RGY (river water-glucose-yeast) medium for the isolation of fungi at 20℃ in the dark. The isolated hyphae were subcultured under the same conditions to observe the morphological characteristics of the colony [6]. The microstructures of the colony and spores were observed after staining with lactophenol cotton blue under a light microscope (DM2500, Leica Microsystems, Wetzlar, Germany). The unrecorded fungus used for observation was deposited at the Korean Collection for Type Cultures (KCTC).
A plant tissue genomic DNA extraction kit (Xi’an Tianlong Science & Technology, Shaanxi, Taiwan) was used to extract the genomic DNA of this fungus. The extracted genomic DNA was selectively amplified from internal transcribed spacer ITS1 to ITS2 using ITS1F and ITS4 primers and 28S ribosomal DNA including the D2 region using LROR and LR5 primers. The selective polymerase chain reaction (PCR) amplifications were conducted following the procedures of previous studies [7,8]. The final PCR products were checked by electrophoresis on 1.5% agarose gel, and the nucleotide sequences were analyzed at Cosmo Genetech (Daejeon, Korea). The analyzed nucleotide sequences were compared by BLAST (Basic Local Alignment Search Tool) from NCBI (https://www.ncbi.nlm.nih.gov/) to select the species having the highest sequence similarity. Based on the results, a neighbor-joining tree was made using the MEGA X program with a Kimura-2 parameter distance model and 1,000 bootstrap analysis [9].
Galactomyces pseudocandidus de Hoog & M.T. Sm., Studies in Mycology 50 (2): 503 (2004)
[MB#369187]
The colony diameter after 7 days was about 39.7-58.2 mm on maltose extract agar (MEA) and 34.5-37.1 mm on potato dextrose agar (PDA). The density of the mycelium alternated between dense and sparse as the concentric circles expanded. The surface color was gambogeish gray (Munsell color notation: 7.5Y 8/2) in MEA and light brownish gray (Munsell color notation: 5Y 9/2) in PDA. It had a finely hairy texture similar to that of a chick's feather with a regular margin, having the outermost hyphae rotated clockwise in a finely hairy shape, and there was no exudate. The reverse side was light amberish gray (Munsell color notation: 10Y 8/2) in MEA and light gambogeish gray (Munsell color notation: 5GY 8/2) in PDA [10]. Conidia, measuring about 6.70-9.64×4.01-6.70 μm (n=20), were square to oblong with a hyaline well with lactophenol cotton blue, and a large number of spores were generated as the hyphae were segmented and showed no septa inside (Fig. 1 and Table 1).
Table 1. Morphological characteristics of Galactomyces pseudocandidus NIE34005 isolated from digestive tract of Procambarus clarkii.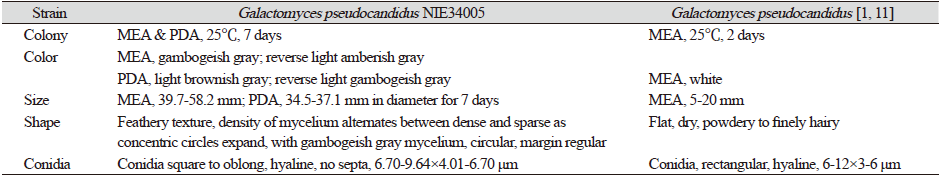
|
|
MEA: Maltose extract agar; PDA: Potato dextrose aga.r |
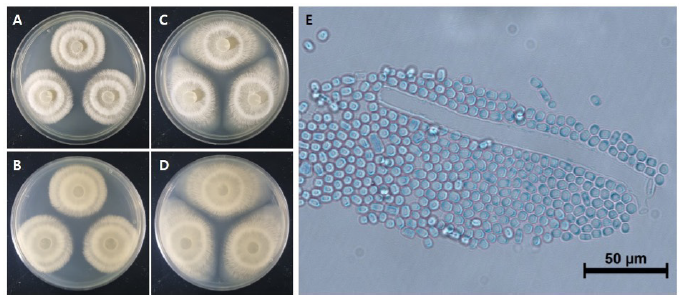
Fig. 1. Cultural characteristics of Galactomyces pseudocandidus NIE34005 isolated from digestive tract of Procambarus clarkii. A, B: Front and reverse sides of colony on maltose extract agar (MEA). C, D: Front and reverse sides of colony on potato dextrose agar (PDA). E: Conidia. Scale bars: E=50 μm.
Specimen examined: Mosan reservoir, Hampyeong-gun, Jeollanam-do, Korea, 35°10'10.16''N, 126°36'7.38''E, 2021.4.7., isolated from the digestive tract of P. clarkii, strain NIE34005, KCTC no. 56818, GenBank no. OK067366 (ITS) and OK067367 (large subunit [LSU]).
Note: A total of five species of the genus Galactomyces have been reported so far. Mainly Galactomyces spp. isolated from soil have been reported. This G. pseudocandidus was the first to be isolated from the stomach of elk (Rucervus eldii Thomas) in Europe [2]. In this study, as G. pseudocandidus was isolated from American crayfish, it is thought that the fungus present in the soil might move to the intestine during the feeding process of animals. The nucleotide sequence from the ITS1 to the D2 region of 28S ribosomal DNA was analyzed and used for the molecular identification of G. pseudocandidus (ITS, KY1034589.1; LSU, MZ412711.1), and each sequence showed 99% homology with the reference sequence of G. pseudocandidus (Fig. 2). G. pseudocandidus has been found in Austria, England, France, Iraq, Netherlands, New Zealand, South Africa, Switzerland, and Turkey so far. Moreover, it has been reported in various environments, such as beet (Beta vulgaris L.), stomach of elk, bark of beech log (Fagus sylvatica L.), sewage rectification filter, and soil, to date [2]. Through this study, G. pseudocandidus, which has not been found in other regions of Asia, isolated from P. clarkii was reported for the first time. Further studies of G. pseudocandidus are needed on the distribution environment of this fungus and fungal function in the animal intestine from an ecological point of view.
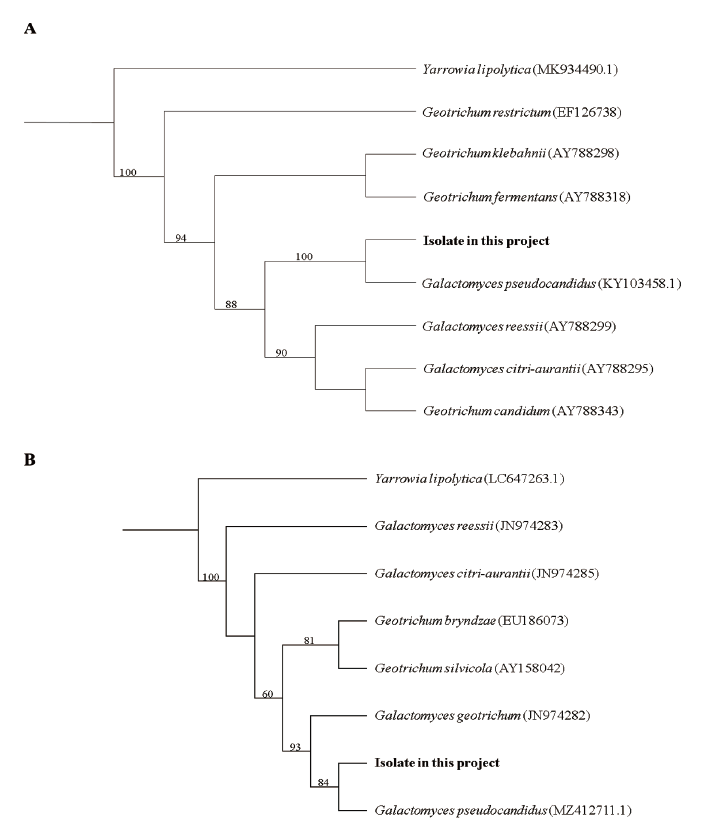
Fig. 2. Phylogenic tree of Galactomyces pseudocandidus NIE34005 isolated from digestive tract of Procambarus clarkii. (A) Internal transcribed spacer (ITS) region, including 5.8Sr ibosomal DNA, and (B) 28S ribosomal DNA were used for the sequence analysis to confirm the topological appropriation of the fungal isolates. Yarrowia lipolytica was used as an out-group, and bootstrap values (>50%) are shown at the branches (1,000 replicates).


