Abstract
In general, mycoviruses remain latent and rarely cause visible symptoms in fungal hosts; however, some viral infections have demonstrated abnormal mycelial growth and fruiting body development in commercial macrofungi, including
Figures & Tables
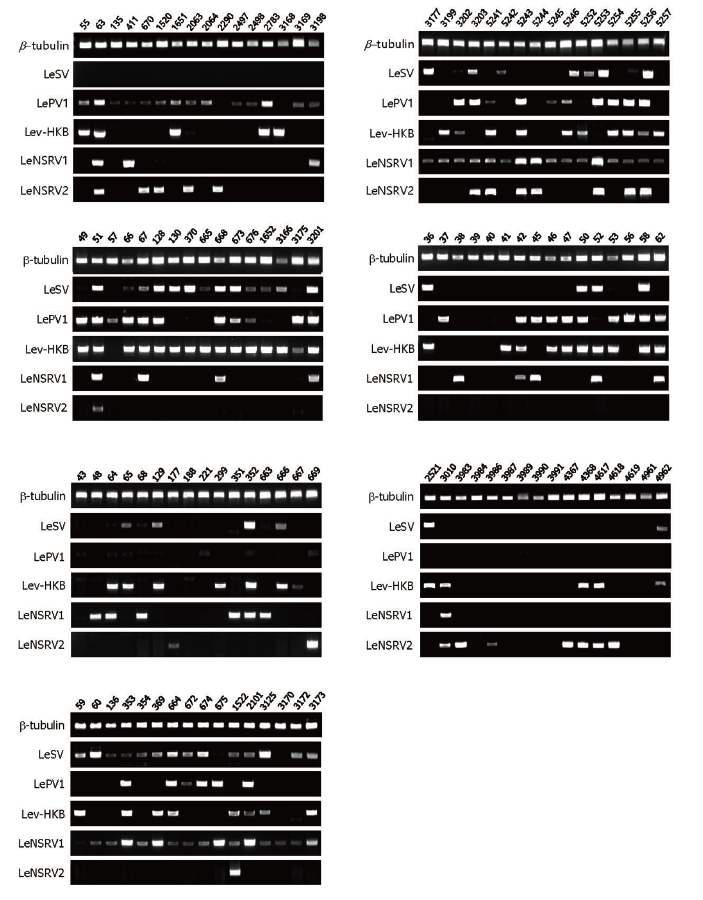
Fig. 1. Reverse transcription-polymerase chain reaction (RT-PCR) detection of viral infection from 112 wild strains of . Specific primer sets targeting RNAdependent RNA polymerase genes of each RNA mycovirus were adopted for RT-PCR. Numbers represent each of the wild strains. Amplification of the β -tubulin gene was used as the positive control to confirm the RNA extraction.


