Introduction
Most Glomeromycotan fungi produce asexual spores in the soil. When their hyphae makes contact with plant roots, they penetrate inside of the root cells and form a mutualistic relationship with the host plant, known as arbuscular mycorrhiza (AM) [1]. Identification of fungi belonging to Glomeromycota has been mainly conducted using morphological characteristics of the spores, including Spore wall structures, ornaments, color, size, and subtending hyphae [2,3], and the molecular analysis. The new Glomeromycota species are continuously being described based on the morphological and molecular evidence of the fungal spores. Approximately 300 Glomeromycota species have been recorded, including 58 species of Acaulospora [4,5].
Genus Acaulospora Gerd. & Trappe (Acaulosporaceae, Diversisporales, Glomeromycetes) was first recorded in 1974 [6]. The spores of Acaulospora are generated initially from the sporiferous saccule [7]. The type of spore formed through this developmental pattern is called as “Acaulosporoid” [7]. Initially, all acaulosporoid species had been located in Acaulospora, and species that form allogeneic spores or showed clearly distinguished morphology have been shifted to other taxa, such as Entrophospora, Kuklospora, or Archaeospora [8,9]. However, Kaonongbua et al. [10] redefined Acaulospora through molecular phylogenetic analysis, and they suggested that Acaulospora can also produce spores other than acaulosporoid. Thus, in the identification of the genus Acaulospora, not only morphological characteristics but also molecular phylogenetic analysis is important.
In Korea, approximately 100 species in Glomeromycota have been reported, including 24 species of Acaulospora [11-14]. However, A. koreana was the only species recorded as a new species of Glomeromycota in Korea so far [14]. In this study, spores of the Glomeromycota were collected in the rhizospheres soil of Miscanthus sinensis from Jeju Island, Korea. Morphological and molecular phylogenetic analyses confirmed that the spores were previously undescribed Acaulospora species. The morphological characteristics and the result of phylogenetic analysis of the new species A. jejuensis were described.
Materials and Methods
The soil samples were collected from Jeju Island of Korea between April and May 2018. Rhizosphere soils were collected around the roots of various host plants. The soil samples were placed in polyethylene bags and transported to the laboratory within 48 hours. For the proliferation of spores, the collected field soils were mixed with autoclaved sand (1:1 volume). Sorghum bicolor as a host plant was planted in a plastic pot [12] and cultivated for about four months in a greenhouse where it was watered once a day. After cultivation, the soils were harvested and used to isolate fungal spore.
The fungal spores were isolated from the soil using wet sieving and sucrose density gradient centrifugation methods [15]. After mounting spores in polyvinyl-lacto-glycerol (PVLG) and Melzer’s reagent, morphological characteristics of spores, such as color, size, wall structures, and surface ornaments of spore walls under optical microscope (AXIO Imager A1, Carl Zeiss, Oberkochen, Germany). Voucher specimens were deposited at the Mycology Lab, Korea National University of Education, Cheongju, Korea.
Nested polymerase chain reaction (PCR) was performed for molecular identification. SSU-ITS-LSU fragment DNA, including the partial small subunit rDNA (SSU), the entire internal transcribed spacer (ITS) region, including 5.8S rDNA, and both the D1 and D2 regions of large subunit rDNA (LSU) [16] were used for amplification. A spore was placed in a 0.2 mL tube with 2 μL sterile water. The genomic DNA was extracted by crushing the spore. The primers used in the reaction were all adjusted to a concentration of 10 ρmol before use and then mixed at the same ratio. For the first PCR, 10 μL 2 × PCR smart mix (SolGent Co., Ltd., Daejeon, Korea) and 1 μL each for SSUmAf/LSUmAr primers [16] were placed into a 0.2 mL tube with genomic DNA, and 6 μL sterile water was added to make a final volume of 20 μL. PCR was conducted in a thermal cycler (Applied Biosystems, Massachusetts, USA). The first PCR product was diluted by 1/10, and used as the template DNA for the second PCR. The second PCR was conducted using the same method as the first PCR using the SSUmCf/LSUmBr primer set [16]. After PCR was completed, electrophoresis was conducted on agarose gel for 20 min to confirm a nucleotide sequence fragment about 1,500 bp size, and DNA sequencing was requested (SolGent Co., Ltd., Daejeon, Korea). The analyzed nucleotide sequence was matched using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) to select the species with the highest similarity. A phylogenetic tree was created by the maximum-likelihood (ML) method using the Kimura 2 parameter (K2P) substitution model in MEGA 7 [17]. Ambiguous characters were excluded, and gaps were treated as missing data. Bootstrap replicates (1,000) were computed for the best-scoring ML tree. We conducted Bayesian analysis [18] for calculating posterior probabilities ( >50% majority rule consensus trees) on the phylogenetic tree branches.
Results
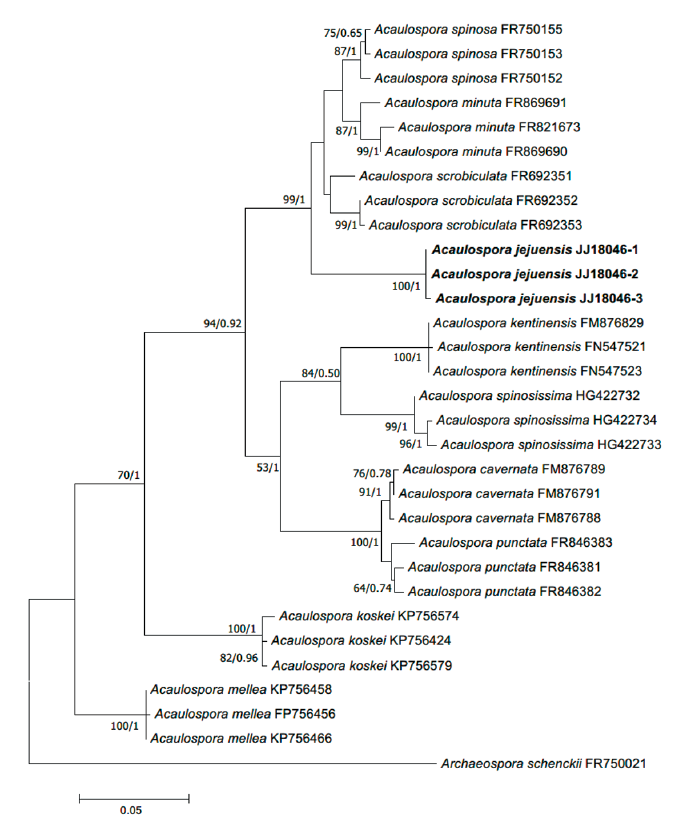
Fig. 1. Maximum-likelihood phylogenetic tree of Acaulospora jejuensis (strain JJ18046-1, 2, 3) based on the alignment of SSU-ITS-LSU DNA sequences. Bootstrap values and Bayesian posterior probabilities are indicated below branches. Archaeospora schenckii was used as an outgroup. The strains used in this study are in bold.
Acaulospora jejuensis H. Park & A. H. Eom, sp. nov. Fig. 1
Mycobank No. MB841896
Type: Korea, Jeju-do, Jeju-si, N33°26'17.0", E126°39'16.2", in rhizosphere soils of Miscanthus sinensis, April 6, 2018, JJ18046 (Holotype), JJ18061 & JJ18094 (isotypes). GenBank no. MN784845 (holotype).
Etymology: The location where the species was first discovered.
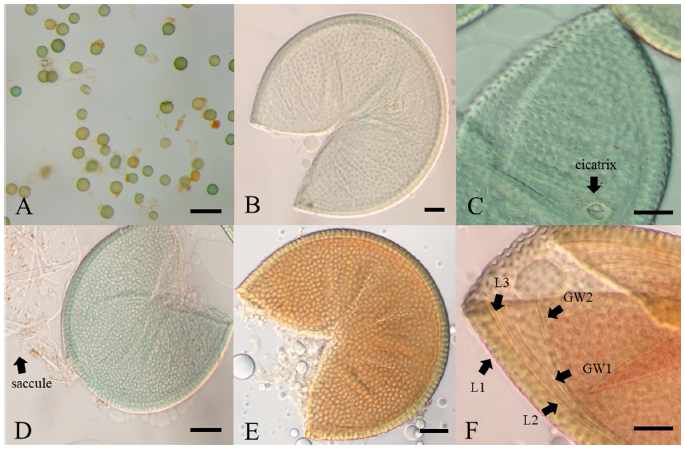
Description: Sporocarps unknown. Spores form solely from the sporiferous saccule in soil; green or turquoise, rarely yellowish–green (Fig. 1A); globose or subglobose, (142-) 189 (-244) × (146-) 201 (-246) μm (n=200). Spore wall structure is composed of three walls: a Spore wall and two germination walls. Spore wall consists of three layers (L1, L2, and L3). L2 surface ornamented with numerous pits with the form of craters. In the mature spores, the site where the saccule neck was separated can be observed as cicatrix. The L1 layer was flexible with a thickness of about 1.0 µm. The L2 layer was the laminae layer, pale green to yellowish-green, and 4.1-6.8 µm thickness. The L3 layer was associated with the L2 layer, and it showed a very thin hyaline layer with a thickness of less than 1 µm. Germination walls GW1 and GW2 are located inside the Spore wall. GW1 showed 0.5-0.9 µm thickness, and it was easy to separate from the Spore wall. It turns reddish-brown or scarlet in response to Melzer’s reagent. GW2 was very close to GW1 and showed 0.8-1.0 µm thickness. The pit-shaped ornaments on the surface of the L1 layer were polygonal or irregularly ellipsoid, and their size ranged from 1.3-2.4 µm in diameter.
Phylogenetic analysis:BLAST results, the SSU-ITS-LSU sequence of A. jejuensis showed 94.26% similarity with Acaulospora scrobiculata MT765465.1 and 94.70% similarity with Acaulospora spinosa FR750155.1. However, the phylogenetic analyses using the maximum likelihood methods and Bayesian methods placed A. jejuensis in a separate clade within Acaulospora (Fig. 2).
Discussion
The morphological characteristics of the fungal spores of JJ18046 were similar to A. minuta, A. scrobicultata, or A. spinosa. JJ18046 showed the numerous pitted ornaments on the surface of spore, which is the morphological characteristic that these 3 species have in common. However, JJ18046 showed several morphological differences with these species. When JJ18046 was compared with A. minuta, it was confirmed that the diameter of pitted ornamentation was larger than that of A. minuta. The average pit size of JJ18046 was 1.3-2.4 µm, whereas that of A. minuta was 0.5-1.8. [19]. A. minuta mainly forms orange spores [19], whereas JJ18046 formed turquoise or yellowish–green spores. JJ18046 showed three layers of Spore wall and two layers of germination wall (Fig. 2F), however, A. minuta has three layers in middle wall between the outer Spore wall and germination wall [19]. The average spore size of JJ18046 (189×201 µm in diam.) was larger than that of A. scrobiculata (120 µm in diam.) [20]. The shape of the pits on the surface of A. scrobiculata was mostly rounded [20], whereas that of JJ18046 was mostly polygonal or irregularly ellipsoidal. Compared to A. spinosa, the spines outside L1 Spore wall, which are the peculiar morphological characteristic of A. spinosa [21], were not observed in the spore of JJ18046. It was also notable that A. spinosa showed yellowish–brown to dark reddish-brown [21] (Table 1).
Table 1. The morphological comparison of Acaulospora jejuensis spores with the other Acaulospora spores.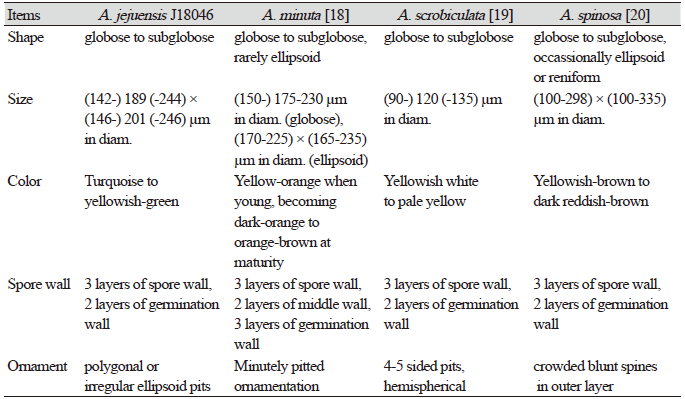
|
In the inferred phylogeny, JJ18046 formed a distinct clade with a strong statistic supports, and shows a close relationship with A. minuta, A. scrobiculata, and A. spinosa (Fig. 1). The formation of pits on the surface of a spore is also a morphological characteristic of species such as A. cavernata or A. punctata [22,23]; however, these species belong to distinctly different clades from JJ18046 on the phylogenetic tree. JJ18046 appears to be the closest to A. scrobiculata in phylogeny but showed <95% similarity to A. scrobiculata on BLAST, and it was considered to form a monophyletic clade distinct from other species. Based on these differences in morphological characteristics and the results of phylogenetic analysis, this species is considered to be a novel species of Glomeromycota that has not been recorded. Therefore, we would like to report this species as A. jejuensis.