Abstract
Macrofungi are valuable resources as novel drug candidates, new biomaterials, and edible materials. Recently, genetic approaches pertaining to macrofungi have been continuously growing for their identification, molecular breeding, and genetic engineering. However, purification and amplification of fungal DNA is challenging because of the rigid cell wall and presence of PCR inhibitory metabolites. Here, we established a direct PCR method to provide a rapid and efficient method for PCR-grade macrofungal DNA preparation applicable to both conventional PCR and real-time PCR. We first optimized the procedure of lysis and PCR using the mycelia of
Figures & Tables
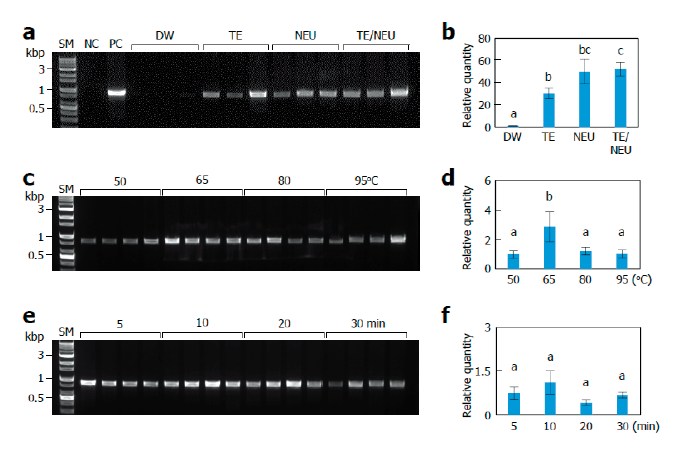
Fig. 1. Effects of lysis conditions. Monokaryotic mycelia of Sanmaru 1ho were lysed with different methods. The resultant lysates were subjected to internal transcribed spacer (ITS) region amplification with conventional PCR (A, C, D) or real-time PCR (B, D, E). The relative quantities of DW, 50℃, and 5 min were set to 1 as reference. The letters, a-c, indicate statistically significant differences based on one-way ANOVA followed by Duncan’s multiple-range test (<0.05). Bars, S.E. (A, B) Effects of lysis solutions. SM, size marker; NC, negative control; PC, positive control; DW, distilled water; TE, Tris/EDTA; NEU, neutralization of KOH with (NH)SO; TE/NEU, neutralization of Tris/EDTA/KOH with (NH)SO. (C, D) Effects of lysis temperatures. (E, F) Effects of heat treatment times.


