INTRODUCTION
Soil fungi play an important ecological role as decomposers, producing a variety of enzymes that break down organic matter, regulating the nutrient balance. They can be important ecosystem regulators and strongly influence plant productivity and diversity [1]. Globally, Ascomycota is one of the largest and most diverse soil-inhabiting fungal taxa, with a relative proportion of 8% corresponding to Sordariomycetes [2]. The class Sordariomycetes (phylum: Ascomycota, subphylum: Pezizomycotina) was introduced by Eriksson & Winka in 1997, and it is the second largest class in the Ascomycota phylum. There is considerable variability among members of Sordariomycetes, in terms of morphology, habitat, and growth form [3-5]. One of the largest genera is Pestalotiopsis, which was first described in 1949 and currently contains 375 species [6]. The morphological characteristics of the genus Pestalotiopsis, which is known to be endogenous to plants and saprotrophic, include conidia with ellipsoidal shape, septate, and appendage-bearing on the apex of the apical cell [7,8]. Recently, Senanayake et al [9]. introduced the family Pestalotiopsidaceae to accommodate seven anamorphic genera, including Pestalotiopsis which possesses pestalotiopsis-like conidia [9]. Pestalotiopsis is distinguished from other genera in this family by the 5-celled, fusiform conidia, with three-colored median cells, hyaline basal cells, and one or more apical appendages. Members of the genus Pestalotiopsis are distributed worldwide, they were mainly found on dead or living plant organs, and some species such as P. terricola, P. humicola, and P. papuana were isolated from soil or other substrates [10]. Several Pestalotiopsis species have been reported to cause serious plant diseases, such as leaf blight in sweet persimmon tree, and canker in Acanthopanax divaricatus [11,12]. In contrast to Pestalotiopsis, another member of the class Sordariomycetes, the genus Botryotrichum comprises only 25 species. This genus belongs to the family Chaetomiaceae and is known to contain saprotrophic fungi. The morphological characteristics of this genus include conidia that are globose to subglobose, solitary, and unicellular [13]. Members of the genus Botryotrichum are commonly found in soils, especially in ones with high levels of organic matter [14]. B. piluliferum has been reported among the soil fungal communities in countries like Canada [15], however, there are still no reports of plant pathogenic species of the genus Botryotrichum.
The aim of this study was to identify unreported fungal species isolated from soil samples in Korea. In order to contribute to enlarging our knowledge about fungal diversity, we present the morphological and molecular characteristics of two species of the class Sordariomycetes, KNUF-21-006 and KNUF-21-028, previously unreported in Korea and now identified as Pestalotiopsis clavata and Botryotrichum iranicum, respectively.
MATERIALS AND METHODS
Sample collection and fungal isolation
Soil samples were collected from Yeongchuk Mountain, Ulju-gun, Gyeongbuk Province (35°30′53.1″N 129°03'08.1″E), and Daro stream, Cheongdo-gun, Hwayang-eup, Gyeongbuk Province (35°39′47.9″N 128°42′58.0″E), Korea. The soil samples were collected between the soil surface and 20 cm below the surface, stored in a sterile plastic bag, brought to the laboratory, and stored at 4℃ until analysis. Serial dilution of the soil samples was performed by weighing 1 g of soil and suspending it in 10 mL of sterile distilled water to prepare serial dilutions (10-1 to 10-5). Each soil serial dilution was vortexed, and 0.1 mL of each dilution was spread onto potato dextrose agar plates (PDA; Difco, Detroit, MI, USA). Well-developed individual colonies were isolated and cultured again on fresh PDA plates and incubated at 25℃ until the mycelium developed. The pure cultures were preserved in 20% glycerol at -80℃ for future studies. Stock cultures of both strains, namely, KNUF-21-006 (NJLNFGC000000047) and KNUF-21-028 (NJLNFGC000000042) were deposited in the National Institute of Biological Resources (NIBR) as metabolically inactive cultures.
Morphological characterization
Morphological characteristics of the isolated strains KNUF-21-006 and KNUF-21-028 were studied by growing them on PDA for 7 days of incubation at 25℃ [16,17]. Cultural characteristics, such as colony color, texture, growth, shape, and size were recorded. The microscopic characteristics were observed using a light microscope (BX-50; Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Fungal strains were grown on PDA plates, and the scraped mycelia were used for genomic DNA extraction using a HiGeneTM Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s instructions. For molecular identification of each strain, target regions, primers, and PCR conditions were based on previous reports [16,17]. The internal transcribed spacer (ITS) region, translation elongation factor 1-alpha (TEF) gene, and beta-tubulin (TUB) gene were used for the identification of strain KNUF-21-006; and ITS, TUB, and large subunit (LSU) rRNA gene were applied for the identification of strain KNUF-21-028. PCR amplification was carried out using the ITS1F and ITS4 primers to amplify the ITS region, NL1 and NL4 primers for LSU, EF1-728F and EF2 for TEF, and TUB amplification was carried out with T1 and Bt2b primers for KNUF-21-006 and T1 and T2 primers for KNUF-21-028 [18-21]. The thermal cycling conditions were as follows: for every primer pair, an initial denaturation step of 95℃ for 5 min and a final extension step of 72℃ for 10 min were used. For ITS 35 amplification cycles of 95℃ for 30 s, 55℃ for 45 s, and 72℃ for 90 s; for TEF 35 cycles of 95℃ for 30 s, 55℃ for 50 s, and 72℃ for 90 s; for LSU 35 cycles of 95℃ for 45 s, 50℃ for 45 s, and 72℃ for 60 s; for TUB 35 cycles of 95℃ for 30 s, 55℃ for 50 s (KNUF-21-006) or 60℃ for 30 s (KNUF-21-0028), and 72℃ for 60 s. The amplified PCR products were purified using EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced at Solgent Co., Ltd. (Daejeon, Korea). Closely related strains were identified using the BLAST search program on the National Center for Biotechnology Information (NCBI) website. The obtained sequences were submitted to the NCBI GenBank database (accession numbers are listed in Table 1).
Phylogenetic analysis
To construct the phylogenetic trees, we analyzed the ITS, TUB, and TEF sequences for strain KNUF-21-006 and ITS, LSU, and TUB sequences for strain KNUF-21-028, retrieving allied sequences from the NCBI GenBank database (Table 1). Sequences were edited using BioEdit v.7.0.5.3 [22] and datasets of the corresponding markers were first analyzed separately and then concatenated into a combined dataset for each strain. Phylogenetic trees were constructed using the maximum likelihood algorithm, with the Kimura model and MEGA software (version 7.0) for 1,000 replications [23].
RESULTS AND DISCUSSION
Pestalotiopsis clavata Maharachch. & K.D. Hyde, Fungal Diversity 56 (1): 108 (2012) [MB#800524] (Fig. 1)
Specimen collection: Yeongchuk Mountain, Ulju-gun, Gyeongbuk Province (35°30′53.1″N 129°03′08.1″E), isolated from soil.
Morphological characteristics of KNUF-21-006
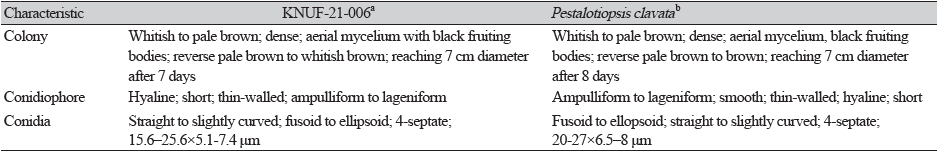
The colonies have dense, aerial mycelium, whitish to pale brown in color, with black fruiting bodies; the reverse of the culture was pale brown to whitish brown. The colony diameter reached 70 mm on PDA after 7 days at 25℃ (Figs. 1A and B). Conidiogenous cells, hyaline, short, thin-walled, discrete ampulliform to lageniform. Conidia straight to slightly curved, fusoid to ellipsoid, four-septate, 15.6–25.6×5.1–7.4 μm (n=20), basal cell hyaline, thin-walled, with conic to obconic with obtuse end. The conidia have three median cells, doliiform, olivaceous to brown; wall rugulose, together 11.5–21.2 μm (n=20) long second cell from base 4.0–6.7 μm (n=20); third cell 3.5–7.5 μm (n=20); fourth cell 4.0–7.0 μm (n=20); apical cell 4.0–6.4 μm (n=20) long; basal appendage mostly 1-3 present, 6.8–18.6 μm (n=20) long (Figs. 1C and D). These morphological characteristics matched well with those typical of the genus Pestalotiopsis and closely agreed with the characteristics of P. clavata [16] (Table 2). The culture and morphological characteristics of the isolated strain KNUF-21-006 suggested that it is most closely related to P. clavata [16].
Molecular phylogeny of the KNUF-21-006
Sequences containing 611, 720, and 483 bp were obtained, which corresponded to the ITS region, TEF, and TUB genes, respectively. A BLAST search of the NCBI database revealed high similarities between the ITS regions of KNUF-21-006 and Pestalotiopsis lushanensis NB3-5 (MW142353; 100%), P. clavata MFLUCC 12-0268 (JX398990; 99.8%), P. pini MEAN 1092 (MT374680; 99.7%), and P. grevilleae CBS 114127 (NR_147548; 99.3%). Based on TUB gene sequence similarities, the closest relatives of the isolate were identified as P. lushanensis NB3-5 (MW147688; 100%), P. pini MEAN 1092 (MT374705; 100%), and P. clavata MFLUCC 12-0268 (JX399025; 99.7%). Based on the TEF gene sequence of KNUF-21-006, 100, 97.2, and 96.5% similarity was observed with the closely related P. clavata MFLUCC 12-0268 (JX399056), P. pini MEAN 1092 (MT374693), and P. lushanensis NB3-5 (MW147695), respectively. Separate analysis of ITS, LSU, or TUB sequences did not allow accurate identification of the novel strain at the species level. Considering that the sequences of ITS, TUB, and TEF are available in GenBank for almost all Pestalotiopsis species and that phylogenetic analysis based on combined ITS, TUB, and TEF sequences allowed the precise differentiation of Pestalotiopsis species investigated in previous studies [16], we also conducted a phylogenetic analysis using concatenated ITS, TUB, and TEF sequences (Table 1). The maximum likelihood phylogenetic tree based on concatenated sequences (Fig. 2) demonstrated that KNUF-21-006 occupies a position within the genus Pestalotiopsis, and the clustering of the isolate with P. clavata indicates that this is the closest relationship at the species level. To our knowledge, this is the first report on this fungal species in Korea.

Fig. 2.Maximum likelihood phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions, beta-tubulin (TUB), and elongation factor (TEF) genes showing the phylogenetic position of strain KNUF-21-006 among Pestalotiopsis species and its closest relationship with Pestalotiopsis clavata . Bootstrap values greater than 50% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold. The tree was rooted using Pseudopestalotiopsis cocos CBS 272.29 as an out-group. Bar, 0.02 substitutions per nucleotide position.
Botryotrichum iranicum A. Alidadi, Mycological Progress 19 (12): 1578 (2020) [MB#831975]
Specimen collection: Daro stream, Cheongdo-gun, Gyeongbuk Province (35°39′47.9″N 128°42′58.0″E), isolated from soil.
Morphological characteristics of KNUF-21-028
The strain KNUF-21-028 was cultured on PDA medium for 7 days at 25℃ to study its cultural and morphological characteristics. The colony diameter reached 24 mm on PDA plates, off-white color, with irregular margins, and canaliculate, dense, smooth aerial mycelium; the back color was flavous (Figs. 3A and B). Conidiophores were septate and hyaline and were produced endobasidially or apically from hyphae. Conidia were globose, single, hyaline, asperate, 8.3–16.8 μm (n=20), and the hilum was clearly visible. No sexual morphs were observed (Figs. 3C and D). These morphological characteristics matched well with those typically described for the genus Botryotrichum and closely agreed with the characteristics of B . iranicum [17] (Table 3). The culture and morphological characteristics of the isolated strain KNUF-21-028 suggested that it was most closely related to B . iranicum [17].
Molecular phylogeny of the KNUF-21-028
From the results of the sequencing analysis, 523, 471, and 650 bp sequences were obtained from the ITS, LSU, and TUB, respectively. A BLAST search of the NCBI database revealed similarities of 99.8% between the ITS regions of KNUF-21-028 and Botryotrichum iranicum ABRIICC 10152 (MN134583), B . iranicum ABRIICC 10153 (MN134584) and B. artogriseum CCF 5752 (LR584032); 100% similarity to B. piluliferum CBS 579.63 (MH858364) was observed. Based on the sequence similarity of the LSU region, the closest relatives of the isolate were identified as B . iranicum ABRIICC 10152 and ABRIICC 10153 (MN134576 and MN134577), B. piluliferum CBS 579.63 (MH869989), and B. verrucosum CBS 116.64 (LT993567), all with 100% similarity. The TUB gene sequence of KNUF-21-028 shared 100% identity with closely related B . iranicum ABRIICC 10152 and ABRIICC 10153 (MN128435 and MN128436), and only 96.4% identity with B. artogriseum CCF 5752 (LR584034). The comparison of ITS, LSU, and TUB loci clearly indicated that isolate KNUF-21-028 belongs to the genus Botryotrichum, but comparative analysis using the sequence of only one gene did not allow precise identification of the novel strain at the species level. Considering that sequences of the ITS, LSU, and TUB regions are available in GenBank for almost all Botryotrichum species, phylogenetic analyses based on combined ITS, LSU, and TUB sequences allowed us to accurately differentiate Botryotrichum species investigated in previous studies [17]. We also conducted a phylogenetic analysis using concatenated ITS, LSU, and TUB sequences (Table 1). The topology of the maximum likelihood phylogenetic tree based on concatenated sequences (Fig. 4) confirmed that KNUF-21-028 belongs to the genus Botryotrichum and showed that this isolate is most closely related to B . iranicum at the species level. To our knowledge, this is the first record of this species in Korea.
Previous studies have reported the isolation of Pestalotiopsis clavata from viable leaves in the Hunan and Yunnan Provinces, China [16]. Some Pestalotiopsis species are of great importance due to their plant pathogenicity [24]. Plant diseases caused by Pestalotiopsis species have been also reported in Korea, including gray blight in tea plant caused by P. theae, leaf blight in sweet persimmons caused by P. diospyri, and canker in Acanthopanax divaricatus caused by P. ellipsospora [12,13,16]. Out of the Pestalotiopsis species isolated in Korea, 11 have been reported as pathogenic [25]. Non-pathogenic Pestalotiopsis species have been found in Korea as endophytic fungi [26], associated with persimmon tree bark [7], in indoor air samples [27] or soil samples [28]. The species-rich genus Pestalotiopsis includes numerous plant pathogens as well as endophytes, which synthesize more than 300 secondary metabolites possessing various biological activities [29]. Seventy new bioactive secondary metabolites have been reported from P. fici including pestalofone F, pestalodiol C, and chloropestolide A showing high cytotoxicity against HeLa and MCF-7 cells [30]. Antifungal pestafolide A and pestaphthalide A were isolated from P. foedans [31] and anti-angiogenic dihydroxanthenone AGI-B4 is produced by P. clavispora [32]. P. clavata is not yet known as the producer of bioactive compounds; therefore, isolate KNUF-21-006 could be considered as the indigenous strain for further study of this species in Korea.

Fig. 3.Maximum likelihood phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions, large subunit (LSU) region, and beta-tubulin (TUB) genes showing the phylogenetic position of strain KNUF-21-028 among Botryotrichum species and its closest relationship with Botryotrichum iranicum . Bootstrap values greater than 50% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold. The tree was rooted using Achaetomium luteum CBS 544.83 as an out-group. Bar, 0.02 substitutions per nucleotide position..
Most Botryotrichum species have been isolated from outdoor environments, such as B . iranicum, first discovered in a soil sample in Iran [17]. There are no reports of plant pathogenicity for representatives of the genus Botryotrichum, which comprises just 25 species. The genus Botryotrichum includes significantly fewer species than Pestalotiopsis, consequently, the number of isolated and identified secondary metabolites from its representatives is not so large. Among them, B. piluliferum is more studied and known as the producer of oxisterigmatocystins E, G, and H displaying antimalarial activity towards Plasmodium falciparum and cytotoxicity against KB, MCF-7, and NCI-H187 cell lines [33], altersolanol C, diorcinol, and botryobutenolide A possessing antibacterial activity against Staphylococcus aureus and Enterococcus faecalis [34].
Although the genus Botryotrichum was first reported in Korea in 1995 with two unidentified strains isolated from traditional Nuruk [35], until now, there are no detailed reports of any Botryotrichum species. Just like P. clavata, B . iranicum has not yet been investigated for the production of bioactive compounds; therefore the isolate KNUF-21-028 can be considered as the indigenous strain for further study of this species in Korea. Overall, further research on Pestalotiopsis and Botryotrichum species is required for a better understanding of their diversity, geographical distribution, pathogenicity, and ecological and biological roles, especially, under Korean environmental conditions.
To our knowledge, this is the first record of the two species, namely, Pestalotiopsis clavata KNUF-21-006 and Botryotrichum iranicum KNUF-21-028, in Korea.