Introduction
Penicillium is a genus of Ascomycetous fungi belonging to the Aspergillaceae family, which plays an important role in the environment, and food and drug production [1]. Penicillium is a large and diverse genus that currently comprises 354 known species [2]. It has a cosmopolitan distribution and huge economic impact on human life [3]. Its main role in nature is decomposing organic materials, such as the devastating rotting caused by pathogenic species in pre- and post-harvest food crops [3]. Some species have positive effects on the food industry, such as in the production of cheeses, like Camembert, and fermented sausages [4].
Morphological identification is the traditional method used for the identification of Penicillium species [5]. Recently, along with morphological identification, molecular methods are being used extensively to study phylogenetic relationships among Penicillium species [6, 7]. In addition, fungal species can be identified using morphological, biochemical, and molecular markers [8]. Molecular tools are required for the identification of Penicillium species, but the choice of accurate markers for use in the genus is challenging [8]. rDNA internal transcribed spacer (ITS), β-tubulin (tub2/BenA), and calmodulin (CaM) are the most commonly used markers for the identification of fungal species [2]. Using these morphological and molecular characterization tools, we report for the first time new P. pimiteouiense fungal isolate from soil samples in South Korea. P. pimiteouiense was first isolated from polycystic kidney cultures [9], and has also been isolated from sandy beach soil samples in Penang Island, Peninsular Malaysia [10]. This study aimed to (i) identify the Penicillium isolates using morphological and molecular characteristics, (ii) use a precise molecular identification tool to compare between the species of Penicillium, and (iii) compare and contrast between the previously reported species and our newly identified species.
Materials and Methods
Soil sampling and isolation of fungi
Soil samples were collected from agricultural fields at various locations in Gyeongsangbuk-do (35o33'03.95''N, 128o28'27.91''E), Korea, in 2017. Soil samples were collected from a depth of 0-15 cm, air dried, and stored in plastic bags at 4℃ until further use. Fungi were isolated using a conventional dilution plating technique [12] and cultured on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) supplemented with 100 μg/L chloramphenicol (a bacteriostatic agent) for 5-7 days at 25℃ until fungal colony growth was observed. The pure cultures were preserved on PDA slants at 4℃ for future use.
Morphological characterization
The morphological characteristics of the isolate KNU17-56 was observed on PDA (MB Cell, Los Angeles, CA, USA), oatmeal agar (OMA; MB Cell), malt extract agar (MEA), Czapek yeast extract agar (CYEA), and yeast extract sucrose agar (YESA). All media were prepared as previously described [12]. The strains were inoculated at three points on 9-cm petri dishes and incubated at 25℃ in the dark for 7 days. After incubation, the diameter of the colonies on various agar media was measured, and the degree of sporulation was determined. Colony color (obverse and reverse sides) was described based on the guidelines provided by Kornerup and Wanscher [13]. Photomicrographs were taken using an HK 3.1 CMOS digital camera (KOPTIC Korea Optics, Seoul, Korea) attached to an Olympus BX50F-3 microscope (Olympus Optical Co., Ltd., Tokyo, Japan) and a scanning electron microscope (LEO Model 1450VP Variable Pressure Scanning Electron Microscope, Carl Zeiss, Cambridge, MA, USA).
Genomic DNA extraction, PCR amplification, sequencing, and data analysis
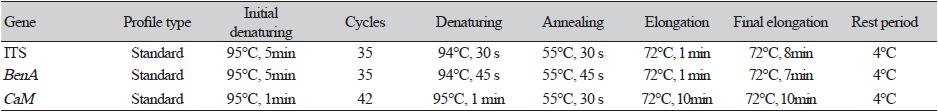
Fungal DNA was extracted from KNU17-56 using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The ITS, BenA, and CaM gene sequences were amplified using the following primers: ITS1 (5ʹ- TCCGTAGGTGAACCTGCG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) [14]; Bt2a (5ʹ-GGTAACCAAATCGGTGCTTTC-3ʹ) and Bt2b (5ʹ-ACCCTCAGTGTAGTGACCCTT-3ʹ) [15]; and CMD5 (5ʹ-CCGAGTACAAGGCCTTC-3ʹ) and CMD6 (5ʹ-CCGATAGAGGTCATAACGTGG-3ʹ) [16]. The amplicons were sequenced and then analyzed using an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Details of the PCR used for amplification and sequencing are listed in Table 1. The sequences were compared with reference ITS, BenA, and CaM gene sequences from GenBank at the National Center for Biotechnology Information (NCBI) using the basic local alignment search tool [17]. Nucleotide sequences of the isolate was deposited in the culture collection of the National Institute of Biological Resources (NIBR, Incheon, Korea). The NIBR numbers was NIBRFG0000501884. The nucleotide sequences were also deposited in GenBank and assigned the accession number MH231760 for KNU17-56. ITS, β-tubulin (BenA) and Calmodulin (CaM) gene sequences of Penicillium species and their strains were used to construct a phylogenetic tree for KNU17-56. GenBank accession numbers are given in Table 2. Phylogenetic relationships were analyzed using the molecular evolutionary genetic analysis (MEGA 6) software [18]. A neighbor-joining tree was constructed using the Kimura 2-parameter substitution model [19]. Bootstrap analysis was performed with 1,000 replications to determine the support for each clade.
Results
Morphology of fungal isolate KNU17-56
Macromorphology of KNU17-56
Photomicrographs of the morphological structures of KNU17-56 are shown in Fig. 1. On PDA, the colonies grew moderately and reached a diameter of 40-45 mm after 7 days at 25℃. The front side of the mycelium had a white margin (6 mm) and a creamy white center, while the back side of the colony had a white margin (5 mm) at the edge and red center (Figs. 1A and 1F). The colony sporulation was dense, conidia were in mass, surface was smooth, texture was floccose, form was circular, elevation and entire margin was flat, and exudate was absent. On CYEA, the colonies grew slowly and reached a diameter of 20-25 mm after 7 days at 25℃. The front side of the colony was white and back side was light yellow (Figs. 1B and 1G). The colony sporulation was moderate to dense, conidia were in mass, surface was smooth, texture was floccose, form was circular, elevation and entire margin was raised, and exudate was absent (Figs. 1B and 1G). A woolly appearance was observed on the front side of the colony (Figs. 1B and 1G). On MEA, the colonies grew moderately and reached a diameter of 25-30 mm after 7 days at 25℃. Front side of the colony was white and back side was light yellow (Figs. 1C and 1H). The colony texture was floccose, form was circular, elevation and entire margin was flat (Figs. 1C and 1H). Sporulation was moderate to dense, surface was smooth, exudate was absent, and conidia were in mass. On YESA, the colonies grew moderately and reached a diameter of 22-27 mm after 7 days at 25℃. Front side of the colony was white and back side was pale yellow (Figs. 1D and 1I). The colony sporulation was moderate to dense, exudates were absent, form was circular form, elevation and entire margin was flat (Figs. 1D and 1I), surface was smooth, conidia were in mass. On OMA, the colonies grew rapidly and reached a diameter of 35-40 mm after 7 days at 25℃. Front side of the colony was powdery white and back side was pale yellow (Figs. 1E and 1J). The colony sporulation was dense, texture was floccose, surface was smooth, exudate was absent, form was circular, elevation and entire margin was flat, and conidia were in mass.
Micromorphology of KNU17-56
The conidiophores were monoverticillate and approximately 7-32 μm in length. Penicilli was monoverticillate. The stipes was short (8-12×1.8-2.1 μm) (Fig. 1K). Phialides were ampuliform (4.6-5.7×1.4-3.1 μm; (Table 3). The conidia were present in short chains, globose to subglobose, finely roughened, and 2.0-3.1 μm in size (Fig. 1M and 1N).

Fig. 1.Morphological characteristics of Penicillium pimiteouiense (KNU17-56) grown for 7 days on potato dextrose agar (PDA), czapek yeast extract agar (CYEA), malt extract agar (MEA), yeast extract sucrose agar (YESA), and oatmeal agar (OMA), at 25℃. Front colony from left to right (A-E) and back colony from left to right (F-J) grown on PDA, CYEA, MEA, YESA, and OMA. Conidiophores and conidia images taken by simple microscope (K, L). Scanning electron microscope images of conidiophore (M) and conidia (N) (scale bars: K, M, N=10 μm, L=2 μm).
Molecular phylogeny of the fungal isolates
Molecular phylogeny of KNU17-56
The ITS, BenA, and CaM gene sequences were used to study and compare the phylogenetic relationships between KNU17-56 and previously described Penicillium species. 18S-ITS1-5.8S-ITS2-28S rDNA sequences of KNU17-56 most closely related to P. pimiteouiense (NR121258.1), forming a monophyletic clade group with a bootstrap value of 99% (Fig. 2). In addition, tub2/BenA gene sequences of KNU17-56 were compared with the sequences deposited in the NCBI database, and KNU17-56 showed a close resemblance to P. pimiteouiense (KC344994.1) (Fig. 3), with a bootstrap value of 99%. Moreover, for further confirmation, the CaM gene sequence of KNU17-56 was compared with the CaM gene sequences of previously reported P. pimiteouiense in the NCBI database. The results obtained from the neighbor-joining phylogenetic tree suggested that KNU17-56 closely resembled P. pimiteouiense (HQ646580.1), forming a monophyletic clade with a bootstrap value of 100%. (Fig. 4).

Fig. 2.Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequences of Penicillium pimiteouiense (KNU17-56) obtained from field soil samples in South Korea. A phylogenetic tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates type strain. Numerical values (>50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000-replicate analysis. Scale bar represents the number of substitutions per site.

Fig. 3.Neighbor-joining phylogenetic analysis of β-tubulin (BenA ) gene sequences of Penicillium pimiteouiense (KNU17-56) obtained from field soil samples in South Korea. A phylogenetic tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates type strain. Numerical values (>50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000-replicate analysis. Scale bar represents the number of substitutions per site.

Fig. 4.Neighbor-joining phylogenetic analysis of Calmodulin (CaM ) gene sequences of Penicillium pimiteouiense (KNU17-56) obtained from field soil samples in South Korea. A phylogenetic tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates type strain. Numerical values (>50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000-replicate analysis. Scale bar represents the number of substitutions per site.
Discussion
Morphological and molecular characterizations are the most commonly used scientific tools for fungal species identification. Mycologists have traditionally used the morphology of fungal species, such as spore producing structures formed from sexual and asexual reproduction, to identify the species [20]. In recent years, morphological identification techniques in combination with molecular techniques are used for species identification within the mycological community [21]. In this study, we characterized our study isolate KNU17-56 as P. pimiteouiense based on their morphological characteristics. The use of differential media such as MEA and CYEA is a simple, easy, and reliable method for identifying Penicillium species [22]. We used five different media (PDA, CYEA, YESA, MEA, and OMA) for the morphological identification and characterization of our study isolate. KNU17-56, isolated from the field soil in Gyeongsangbuk-do province of Korea, was likely to be P. pimiteouiense based on the shape, size, and structure of conidiophores, conidia, phialides, and stipes. The macromorphology and micromorphology of the isolate KNU17-56 matched that of P. pimiteouiense, as previously reported [9, 10]. KNU17-56 only has slight differences from the original description made in previous reports. P. pimiteouiense identified in our study have not been reported in Korea before, therefore, herein is the first report isolated from soil samples in Korea.
The identification of fungal species using molecular tools is a widely used technique by mycologists and taxonomists. DNA sequencing is one of the most reliable tools as it displays the relationship within the species and also facilitates sequence-based identification [2]. rDNA ITS is the most widely used molecular marker in fungi identification [23]. In this study, we used 18S-ITS1-5.8S-ITS2-28S rDNA for the identification of KNU17-56 our results confirmed that our study isolates belong to P. pimiteouiense (Fig. 2). P. pimiteouiense lies near the clade of P. stratisporum when using ITS rDNA for identification [9]. This was also observed in our study (Fig. 2). Therefore, because of this limitation associated with using ITS as a species marker for Penicillium, tub2/BenA was considered the best option for a more precise identification [2, 24]. BenA gene maker analysis showed that KNU17-56 had 99% similarity with P. pimiteouiense (Fig. 3). Another possible secondary marker for Penicillium species identification is CaM [2], which has proven to be a powerful tool for the molecular exploration of taxa related to Penicillium [25, 26]. Phylogenetic analysis using the CaM gene further confirmed that KNU17-56 belongs to P. pimiteouiense (Fig. 4). Some members of the genus Penicillium are responsible for the production of penicillin, which kills or inhibits the growth of some bacteria [27]. Penicillium has a wider application in biotechnology, in the production of enzymes, vitamins, valuable chemicals, and proteins, due to its low cost [28]. Although it also produces mycotoxins which are responsible for food spoilage, the economic importance supersedes the negative effects [28].
In conclusion, this is the first report of P. pimiteouiense from field soil samples in South Korea, identified based on its morphological and molecular characteristics. This fungal isolate may be of biotechnological importance in the field of mycology, therefore, further studies on their use and implications are warranted.
Acknowledgements
This work was supported by a grant from the National Institute of Biological Resources (NIBR) grant funded by the Ministry of Environment (MOE) of the Republic of Korea under the project of survey and discovery of indigenous Korean fungal species in 2017, and a grant from the University Industry Cooperation Foundation of Kangwon National University (KNU).