Introduction
Endophytic fungi are organisms that have a symbiotic relationship with plants and do not cause disease symptoms in their host when balance is maintained [1]. They are found in all plant tissues, such as roots, stems, and leaves, and are transmitted vertically or horizontally [2]. Endophytic fungi provide benefits to their host plants, such as tolerance to water stress [3], increased photosynthetic efficiency [4], and resistance to pathogens [5].
The diversity of endophytic fungi in symbiosis with conifers is affected by the host plant species and the geographic region [6]. In previous studies, endophytic fungi including the genus Lophodermium were found to be common endophytes in conifer species [6-8], and their close relationship with this host plant have been reported [9,10]. In this study, endophytes were isolated from the leaves of two conifer species (Pinus densiflora and Juniperus rigida) during a survey of the diversity of endophytic fungi in Korea. We identified two fungal species that were previously unrecorded in Korea and carried out their morphological and molecular characterization in the present study.
Materials and Methods
Sampling, isolation and morphological characterization
Leaves of Pinus densiflora and Juniperus rigida were collected from Mt. Baegasan, Hwasungun, Jeollanamdo (35°41′52.0″N, 129°21′04.2″E) and Gyeongjusi, Gyeongsangbukdo (35°9′55.93″N, 127°10′18.38″E) in Korea, respectively. The samples were transported to the laboratory within 24 hours for the isolation of endophytic fungi. Randomly selected asymptomatic leaves were washed under running tap water and then surface sterilized through sequential treatment with 30% H2O2 and 70% EtOH for 1 minute each. After washing with sterile water, any remaining liquid on the leaves was removed using sterilized filter paper. After surface sterilization, the leaves were cut to a length 1.5 cm using surface-sterilized scissors and then placed on potato dextrose agar (PDA; Difco Lab., Detroit, USA) and malt extract agar (MEA; Kisan bio, Seoul, Korea) medium. Once the hypha had extended, the endophytic fungi were separated by subculturing on PDA medium. The pure isolated strain was inoculated onto PDA and MEA media and incubated for 7 days at 25℃ in the dark. Conidia formation was induced using the slide culture method, and the hyphae and conidia were observed with an optical microscope.
Molecular characterization
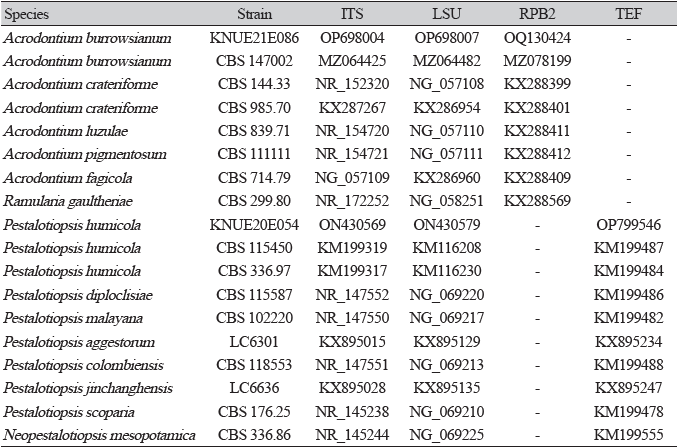
For molecular identification of the species, genomic DNA was extracted using the HiGene™ Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea). The internal transcribed spacer (ITS) and large subunit (LSU) regions of rDNA were amplified using the ITS1F/ITS4 [11] and LROR/LR16 primers [12], respectively. For further sequencing analysis, RNA polymerase II second largest subunit (RPB2) genes in Acrodontium burrowsianum were amplified using fRPB2-5F/fRPB2-7cR primers [13], whereas the translation elongation factor 1-alpha (TEF) gene in Pestalotiopsis humicola was amplified using the EF1-668F/EF1-1251R primers [14]. The PCR products were electrophoresed on a 1.5% agarose gel for 20 minutes. After confirmation of the bands, nucleotide sequence analysis of the genes was conducted by SolGent Co. (Daejeon, Korea). The species similarity of the nucleotide sequences was confirmed using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information, and phylogenetic trees were prepared using the maximum-likelihood method with MEGA11 [15]. The obtained sequences were submitted to the NCBI GenBank database (Table 1).
Results and Discussion
Acrodontium burrowsianum Crous, Persoonia 46: 365 (2021) [MB#839513] (Fig. 1)
Morphological characteristics of strain KNUE21E086
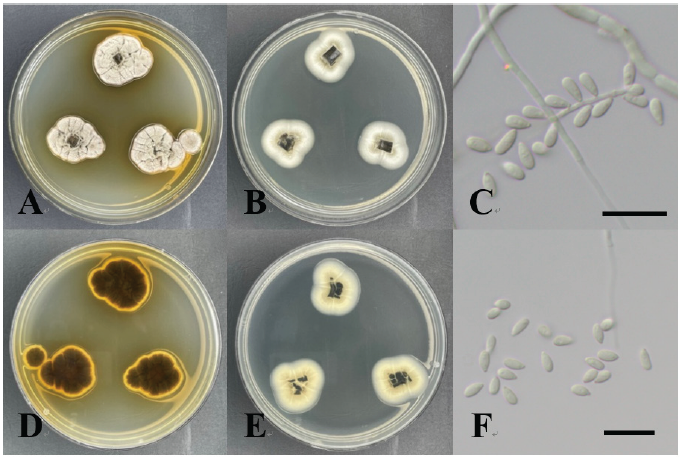
Acrodontium burrowsianum was isolated from the leaves of Juniperus rigida. Colonies cultured for 7 days on PDA medium were 25-30 mm in diameter. The color of the front side of the colony was light brown at the center and changed gradually to a lighter smoke gray at the edge. The reverse side of the colony was dark brown in the center and yellow at the edge. The colony was higher in the central part, with hyphae extending radially, and the margin was irregular. Colonies cultured for 7 days on MEA medium were 20-25 mm in diameter. The front of the colony was smoke gray in color, whereas the reverse side was isabelline or cream colored. The colony was relatively flat in shape and attached to the medium, and the margin was irregular. The conidia, which were smooth and hyaline ellipsoids with an obtuse apex, were attached to the sides of the mycelium. The dimensions of the conidia were (2.64-) 3.79 (-4.65)×(1.44-) 1.92 (-2.25) µm.
Specimen examined: Gyeongjusi, Korea; 35°41′52.0″N, 129°21′04.2″E; May 26, 2021; isolated from the leaves of Juniperus rigida; National Institute of Biological Resources (NIBR) No. NIBRFGC000509082; GenBank No. OP698004 (ITS), OP698007 (LSU), and OQ130424 (RPB2)
Molecular characteristics of strain KNUE21E086
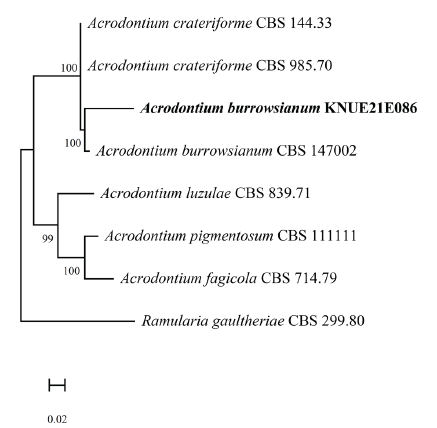
The BLAST results showed that the ITS sequence had 99.08% similarity with that of Acrodontium burrowsianum MZ064425, whereas the LSU sequence showed 99.83% similarity with that of Acrodontium burrowsianum MZ064482. The RPB2 sequence showed 98.97% similarity with the sequence from Acrodontium burrowsianum MZ078199. Maximum-likelihood phylogenetic analysis was performed by combining the ITS, LSU, and RPB2 sequences (Fig. 2). On the basis of its molecular and morphological characteristics, strain KNUE21E086 was identified as Acrodontium burrowsianum.
Pestalotiopsis humicola Maharachch., K.D. Hyde & Crous, Studies in Mycology 79: 165 (2014) [MB#821661] (Fig. 3)
Morphological characteristics of strain KNUE20E054
Colonies on PDA and MEA media were 42-44 and 40-42 mm in diameter, respectively, and had regular margins. White aerial mycelia were observed. The colony was bright ivory in color from the front side and ivory from the reverse side. When cultured on PDA medium, black pycnidial conidiomata were observed. The conidia were brown in color, with a size of (15.59-) 17.53 (-20.76)×(4.81-) 5.03 (-5.53) µm. The 5-cell-shaped conidia had septa and 2-3 tubular apical appendages.
Specimen examined: Mt. Baegasan, Hwasungun, Jeollanamdo, Korea; 35°9′55.93”N, 127°10′18.38”E; May 16, 2020; isolated from the leaves of Pinus densiflora; NIBR No. NIBRFGC000508505; GenBank No. ON430569 (ITS), ON430579 (LSU), and OP799546 (TEF).
Molecular characteristics of strain KNUE20E054
As determined by BLAST analysis, the ITS sequence of strain KNUE20E054 had 99.66% similarity with that of Pestalotiopsis humicola KM199319, and its LSU sequence showed a similarity of 99.68% with that of Pestalotiopsis humicola KM116230. The TEF sequence had 96.23% similarity with that of Pestalotiopsis humicola KM199484. Maximum-likelihood phylogenetic analysis was performed by combining the ITS, LSU, and TEF sequences (Fig. 4). On the basis of its molecular and morphological characteristics, strain KNUE20E054 was identified as Pestalotiopsis humicola.
Acrodontium burrowsianum was first isolated from the leaves of an unidentified Poaceae plant in the Republic of South Africa by Crous et al. [16] in 2021 (Table 2). Although closely related to Acrodontium crateriforme, it is known that Acrodontium burrowsianum has smaller conidia [16]. The conidia of the strain KNUE21E086 reported in this study are smaller than those of Acrodontium crateriforme and similar to the ones of Acrodontium burrowsianum. Pestalotiopsis humicola was first isolated from soil by Maharachch et al. in 2014 [17], and the symbiotic relationship of the fungus with plants was confirmed through that study. Although the conidial morphologies of the genera Truncatella and Pestalotia are similar to that of Pestalotiopsis, these three genera can be distinguished by their differences in the number of cells separated by a septum; that is, Truncatella has 4-cell, Pestalotiopsis has 5-cell, and Pestalotia has 6-cell conidia (Table 3). To the best of our knowledge, this is the first record of the species Acrodontium burrowsianum KNUE21E086 and Pestalotiopsis humicola KNUE20E054 in Korea.