Insects are a remarkably diverse and abundant group of animals on earth [1]. Studies on the insect gut have received considerable attention and have colonized many different habitats [2]. In most insects, gut symbionts are essential for survival and development, suggesting the presence of a core microbiome. Terrestrial insects, such as the soil invertebrates (collembolans, earthworms, and nematodes), have been reported to harbor a rich microbiome and putative symbionts [3]. Earthworms are considered to be excellent bioindicators of the relative health of soil and possess several qualities that predispose them to use in monitoring terrestrial ecosystems worldwide [4]. Earthworms increase microbial activities by providing their gut mucus with energetic and readily metabolizable compounds [5] and considerable physicochemical conditions such as neutral pH, high moisture, and ideal temperature conditions [6]. However, earthworms have poor enzymatic systems and rely on ingested soil microorganisms to degrade soil organic matter [7]. The earthworm gut is an effective tubular structure extending from the mouth to the anus; its different regions include the muscular pharynx, esophagus, intestine, and associated digestive glands. Gut contents usually comprise mucus, organic matter, and mineral matter. An analysis of gut contents in earthworms revealed the presence of different kinds of symbionts such as microfungi, bacteria, and protozoa; most microfungal species are present in the foregut, gradually decreasing in number in the mid and hindgut with the fewest in freshly laid casts [8]. Although the earthworm gut provides ideal conditions for growth of bacterial and fungal colonies [9], interactions between yeasts and earthworms are not well understood. In some associations, the fungus provides digestive enzymes and nutrition to the insect host.
The genus Blastobotrys was originally proposed as a hyphomycete by Von Klopotek [10]. Later, Kurtzman and Robnett [11,12] determined phylogenetic relationships among ascomycetous yeasts and yeast-like taxa based on the phylogenetic analysis of the D1/D2 region of the large subunit (LSU) rRNA gene and demonstrated that the Blastobotrys species are anamorphic members of Saccharomycetales, which are closely related to Arxula and Sympodiomyces and several species of Candida. In 2007, the relationships among species assigned to the ascosporic yeast clades, which include Arxula, Blastobotrys, Sympodiomyces, Trichomonascus, Wickerhamiella, and Zygoascus, were re-examined using phylogenetic analyses of multigene sequences, including the LSU rRNA gene, mitochondrial small subunit (SSU) rRNA gene, and cytochrome oxidase II. Based on multigene phylogenetic analysis, Arxula, Blastobotry, and Sympodiomyces belonged to the Trichomonascus clade, which is the teleomorphic state of the clade. Consequently, the anamorphic genera Arxula, Blastobotrys, and Sympodiomyces, are re-assigned as a single anamorphic genus, with Blastobotrys having taxonomic priority over Sympodiomyces and Arxula [13].
In a biodiversity study on the insect gut microbiome in Korea, unrecorded yeast strains belonging to Rhodotorula taiwanensis, Dirkmeia churashimaensis, Moesziomyces aphidis, Moesziomyces antarcticus, Papiliotrema aspenensis, and Saitozyma flava were isolated [14]. Based on DNA sequence comparisons, strain E4 isolated in Korea has been identified as an ascomycetous yeast species in the gut of the earthworm Eisenia fetida. Sequence analysis of the internal transcribed spacer (ITS) region and D1/D2 domains of the LSU rRNA gene identified this strain as Blastobotrys illinoisensis.
Eight earthworms and soil samples were collected from Gyeonggi-do province (37.4138°N, 127.5183°E), South Korea, and were transported in sterile beakers. Earthworms visually identified as Eisenia fetida were immediately washed with distilled water, frozen in liquid nitrogen, ground using a pestle and mortar, and stored at -80℃ until required. During dissection, frozen earthworms taken out from the freezer and immediately rinsed with 70% ethanol. The digestive tract of each earthworm was extracted by cutting the exoskeletons. The whole gut was stored in 1.5 mL microcentrifuge tubes, dissected, and cut into segments of 0.2-0.4 cm without surface sterilization. Between 30 and 40 tissue segments were then evenly placed in 9 cm diameter onto yeast extract-malt extract (YM) agar plates (pH 3.7-5.0; Difco) supplemented with 0.01% (w/v) chloramphenicol and then incubated at 25℃ in the dark. Colonies appeared after 3-5 days and were transferred to YM broth for propagation. Yeast strains were purified using YM agar and incubated at 25℃. The plates were stored at 4℃, and re-streaking was performed for purification. Purified yeast strains were inoculated in YM broth supplemented with 10% glycerol (v/v) and stored at -80℃. For routine subculturing and maintenance, the strains were grown on YM agar or broth at 28℃.
Strain E4 was characterized morphologically, biochemically, and physiologically according to standard methods described by Kurtzman et al. [15]. Formation of pseudohyphae and true hyphae was investigated via slide culture on potato dextrose agar (PDA; 20% potato infusion, 2% glucose, and 1.5% agar) at 25℃ for 7 days. Ascospore formation was investigated by growing the strain on PDA, corn meal agar, 5% malt extract agar (5% malt extract and 1.5% agar), yeast extract-peptone glucose (YPD) agar (1% yeast extract, 2% peptone, 2% glucose, and 1.5% agar), and YM agar at 15℃ and 25℃ for 1 month. Growth at various temperatures was determined via cultivation on YM agar. Assimilation tests for carbon and nitrogen sources were performed using liquid media. Starved inocula were used in nitrogen and vitamin assimilation tests.
Genomic DNA was extracted and purified using the CTAB method [16]. The ITS (ITS1–5.8 S–ITS2) region of the rRNA gene was amplified using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), as described by White et al. [17]. Amplification of the ITS region was performed using the following conditions: 95℃ for 3 min; followed by 37 cycles of 94℃ for 30 s, 52℃ for 30 s, and 72℃ for 30 s; and a final extension at 7℃ for 10 min. Meanwhile, the D1/D2 region of LSU rRNA was amplified with the primers NL1 (5′-GCATATCAATAAGCGGGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′), as described by Kurtzman and Robnett [12]. Amplification of the D1/D2 region was performed as follows: 94℃ for 6 min; followed by 40 cycles of 94℃ for 60 s, 50℃ for 60 s, and 72℃ for 60 s; and a final extension at 72℃ for 5 min. Sequence alignment was performed using CLUSTAL X 1.83 [18]. Reference sequences were retrieved from GenBank (accession numbers indicated in the phylogenetic tree). Phylogenetic and molecular evolutionary analyses using neighbor-joining [19] and maximum-likelihood methods were conducted in MEGA version 7 [20], and evolutionary distances were calculated using the general time-reversible (GTR) model [21]. Confidence levels of the clades were estimated through bootstrap analysis (1,000 replicates) [22], and only values above 50% were recorded for the resulting trees. Schizosaccharomyces pombe NRRL Y-12796T was used as an outgroup.
Species delineation and phylogenetic placement
A total of 10 yeast strains were isolated from the gut of earthworms during an investigation of earthworm gut-associated microbial communities in South Korea. Sequence analyses of the D1/D2 domains of the LSU rRNA gene revealed that E4 isolate belonged to Blastobotrys illinoisensis. Based on the pairwise sequence similarity of D1/D2 domains, the closest relatives of the species were B. mokoenaii CBS 8435T (99.4%) and B. malaysiensis CBS 10336T (99.2%). The D1/D2 sequences of the unreported species showed a sequence divergence of 0.5% (three nucleotide substitutions and zero gaps over 633 bases), 0.6% (four nucleotide substitutions and no gaps), and 0.8% (five nucleotide substitutions and zero gaps) from the closest relatives, Blastobotrys illinoisensis, B. mokoenaii, and B. malaysiensis, respectively. Results of the pairwise sequence similarity based on ITS regions revealed that the closest relatives of the strain were B. mokoenaii CBS 8435T (95.7%) and B. malaysiensis CBS 10336T (96.6%). A comparison of the ITS sequences revealed that the novel species showed a sequence divergence of 2.5% (14 nucleotide substitutions and 7 gaps over 566 bases), 2.2% (12 nucleotide substitutions and 11 gaps over 539 bases), and 2.1% (12 nucleotide substitutions and 7 gaps over 548 bases) from its closest relatives B. mokoenaii and B. malaysiensis. Furthermore, according to the DNA barcoding analysis of yeast species by Vu et al. [23], ITS and LSU can be used as taxonomic thresholds (96.3 and 99.5%, respectively) to discriminate yeast species. Therefore, we concluded that the novel isolates belonged to Blastobotrys illinoisensis. A phylogenetic tree was constructed using the neighbor-joining method based on the D1/D2 LSU rRNA gene sequences of the unreported species, its closest relatives, and members of the Blastobotrys clade, as defined by Kurtzman et al. [15]. The unreported species formed a clade with B. illinoisensis CBS 10339T (Fig. 1). The phylogenetic relationships of the unreported species and its closely related species in the subclade were also supported by maximum likelihood analysis (data not shown). Schizosaccharomyces pombe NRRL Y-12796T was used as an outgroup.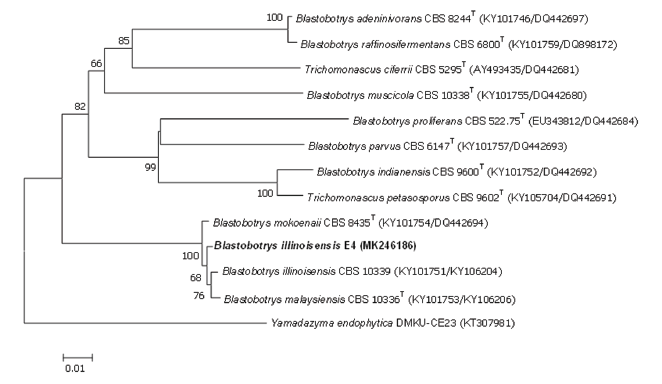
Fig. 1. Phylogenetic placement of Blastobotrys illinoisensis E4 and related species using the neighborjoining method and maximum-likelihood analysis of concatenated sequences of the internal transcribed spacer (ITS) region and the D1/D2 region of the large subunit (LSU) rRNA gene. Yamadazyma endophytica DMKU-CE23 was used as the out-group species. Bootstrap values based on 1,000 replications are shown at the branch nodes. Ba,r 0.01 substitutions per nucleotide position
Collectively, sequence data and phylogenetic analysis of the D1/D2 region of the LSU rRNA (Fig. 2) and the ITS region suggested that the isolate is a novel species of the genus Blastobotrys. Moreover, the ITS region sequence was not available for many Blastobotrys species, except for B. illinoisensis CBS 10339T (DQ898169), B. malaysiensis CBS 10336T (DQ898170), B. raffinosifermentans CBS 6800T (DQ898172), B. adeninivorans CBS 8244T (DQ898168), and B. mokoenaii CBS 8435T (DQ89817).
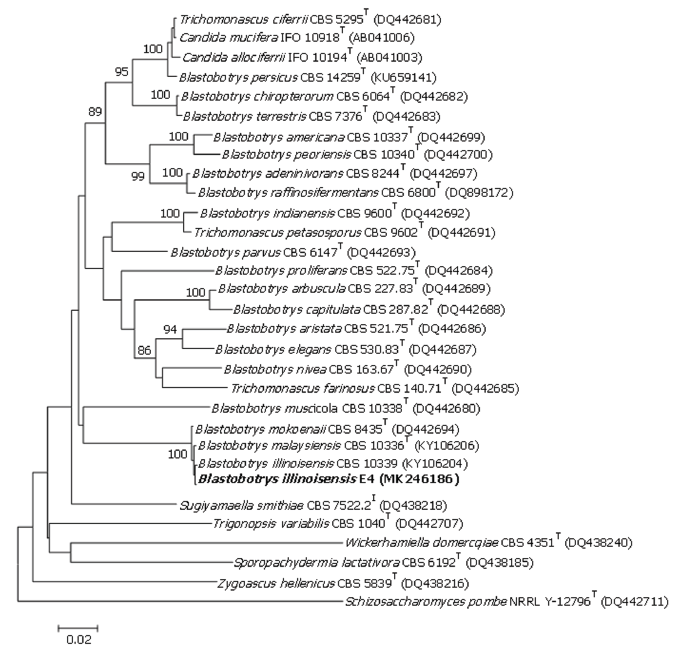
Fig. 2. Phylogenetic placement of Blastobotrys illinoisensis E4 and related species using the neighborjoining method and maximum-likelihood analysis of D1/D2 large subunit (LSU) rRNA gene sequences. Schizosaccharomyces pombe was used as the out-group species. Bootstrap values based on 1,000 replications are shown at the branch nodes. Ba,r 0.02 substitutions per nucleotide position
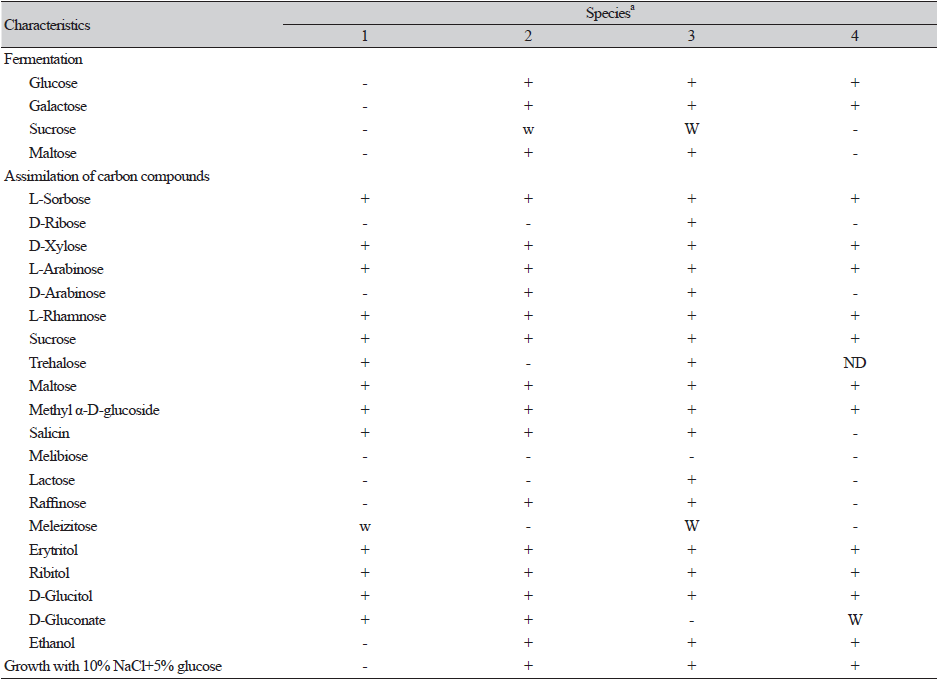
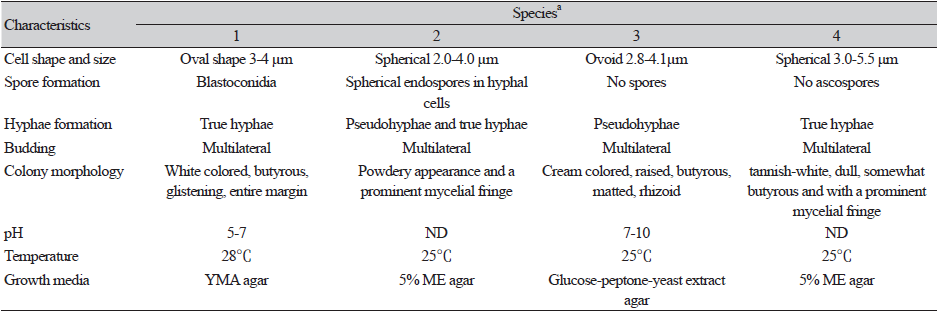
Cells of strain E4 were butyrous, and colonies appeared white and relatively raised. Detailed morphology is provided in the succeeding section. The morphological and physiological characteristics of strain E4 were similar to those of Blastobotrys spp. [24]. Analysis of its sporulation activity revealed the absence of conjugation and formation of asci or ascospores. Budding cells were formed via multilateral budding, and formation of true hyphae was observed (Fig. 3). A similar type of budding has been reported in the closest species Blastobotrys persicus; however, this species lack true hyphae [24]. The detailed phenotypic characteristics of strain E4 and related Blastobotrys spp. are presented in Table 1 and Table 2. In addition, B. koreensis assimilates trehalose and melezitose (weak) and do not assimilate D-arabinose, raffinose, and ethanol, compared to B. illinoisensis. Strain E4 did not ferment carbon sources, did not grow in 10% NaCl containing 5% glucose, and had different assimilation profiles compared to Blastobotrys illinoisensis, B. mokoenaii, and B. malaysiensis (Table 1). Furthermore, strain E4 can be differentiated from the closest relative species based on phenotypic and chemotaxonomic characteristics, and thus the novel species was assigned to the genus Blastobotrys.
Description of Blastobotrys illinoisensis E4
Growth on yeast malt agar (YMA): After incubation for 72 h at 28℃, colonies appear white, butyrous, glistening, and with entire margins (Fig. 3). Cells are spheroidal to ovoidal (3-4×2.5-4 µm), and occur singly, in pairs, or in small clusters. Budding is multilateral (Fig. 2b). True hyphae are observed on YMA and PDA agar. Ascus formation is absent. Pseudohyphae are rare on yeast extract peptone glycerol (YPG) media, PDA, and cornmeal agar. Protuberances, an indication of potential early stage of germ tube production, and short chains of blastoconidia (Fig. 3) are formed. Colonies grow at 15℃ and 37℃, but not at 40℃.
Fermentation is absent. N-acetyl glucosamine, L-arabinose, D-galactose, D-glucose, L-sorbose, D-xylose, L-rhamnose, sucrose, trehalose, maltose, methyl α-D-glucoside, cellobiose, salicin, erythritol, ribitol, D-glucitol, D-mannitol, myo-inositol, D-glucono-1,5-lactone, D-gluconate, D-glucuronate (weak), meleizitose (weak), D-galacturonic acid, glycerol (weak), DL-lactate (weak), succinate, citrate, and xylitol are assimilated. No growth was observed when D-ribose, melibiose, lactose, raffinose, methanol, and ethanol are used. Ammonium sulfate, l-lysine, and cadaverine are assimilated as nitrogen sources. Potassium nitrate, sodium nitrate, ethylamine HCL, and creatine are not assimilated. Growth on 50% and 60% glucose-yeast extract agar is observed, and growth on 0.01 and 0.1% cycloheximide is not observed. No growth was observed in 10% NaCl with 5% glucose. Acid and starch formation tests are negative. Diazonium Blue B (DBB) reaction and urease are negative.
Strain E4 was isolated from the gut of Eisenia fetida collected from Onam-si, Korea. The strain is preserved in a metabolically inactive state at the Korean Collection for Type Cultures (KCTC), Republic of Korea (KCTC 27831) and the Japan Collection of Microorganisms, Japan (JCM 33428). The MycoBank accession number is MB830626.
Acknowledgements
This work was supported by a research grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202231206) 2022 Graduate Program of Undiscovered Taxa; and a grant from the National Research Foundation of Korea (NRF), funded by the Korean government (MSIT) (No.2021R1F1A106138912).
Conflict of interest
All authors certify that there are no conflicts of interest with any financial organization regarding the material discussed in the manuscript.
Ethical approval
This article does not contain any studies involving human participants or animals performed by any of the authors.
Abbreviation
The GenBank/EMBL/DDBJ accession numbers for the combined sequences of the D1/D2 region of the Large subunit (LSU) rRNA gene and the Internal transcribed spacer (ITS) region of strain E4 are MK246186 and MK246187, respectively. The MycoBank number for Blastobotrys illinoisensis is MB830626.