INTRODUCTION
The genus Paraconiothyrium belongs to the family Didymosphaeriaceae, which was introduced from the asexual morphs grouping with Paraphaeosphaeria, and the following four new taxa were described: Paraconiothyrium estuarinum (type species), P. brasiliense, P. cyclothyrioides, and P. fungicola [1]. The species of Paraconiothyrium are polyphyletic within Didymosphaeriaceae and separated from the genera Coniothyrium, Paraphaeosphaeria, Alloconiothyrium, and Dendrothyrium as a paraphyletic group [2,3]. According to the 2022 version of the outline of fungi and fungus-like taxa, Didymosphaeriaceae contains 33 genera and approximately 254 species [4]. Currently, the total 32 described Paraconiothyrium species belong to Didymosphaeriaceae in the Index Fungorum database (https://www.indexfungorum.org/names/ Names.asp: accessed on February 27, 2023). The morphological characteristics of Paraconiothyrium species compromise with the presence of simple or complex conidiomata having a pycnidial or eustromatic type. Conidiogenous cells are discrete or integrated with a percurrent or phialidic nature. Similarly, conidia are primarily aseptate or single septate, thin walled and smooth, and during liberation hyaline, which later turns brown [1]. However, the genus Paraconiothyrium is considered cosmopolitan in nature with diverse host habitats and geographical distributions [5]. Paraconiothyrium species have been most often isolated from warm regions such as Brazil, Italy, Papua New Guinea, South Africa, and Turkey [6]. Similarly, it has also been reported that Paraconiothyrium species are diversely available and found in different sources such as soil, pathogenic fungus, plant endophytic fungi, and marine [7].
The objective of this study was to screen the native fungal species from soil in Korea. Based on morphological and cultural characteristics along with their molecular phylogenetic analysis, the isolated fungus was an unreported species of the genus Paraconiothyrium. Hence, to our knowledge, this is the first report of its isolation and identification in Korea.
MATERIAL AND METHODS
Sample collection and fungal isolation
In 2021, soil samples were collected from Chungcheongbuk Province (37°01ʹ37ʹN, 127°16ʹ45ʺE), Korea, using a sterile spatula and stored in an air-dried plastic bag at 4℃. The isolation was performed using the dilution plating technique, as described previously [8]. A soil sample (1 g) was added to 10 mL of sterile distilled water, and the soil suspension was diluted serially and spread on potato dextrose agar plates (PDA; Difco, Detroit, MI, USA). The plates were incubated for 3-4 days at 25℃ until single colonies were observed. Next, the single colonies were transferred to new PDA plates and incubated at 25℃. The strain KNUF-21-66Q1 was selected based on cultural, morphological, and phylogenetic analyses. The fungal strain was maintained in 20% glycerol at -80℃ for further investigation. Stock cultures of the strain KNUF21-66Q1 (NIBRFGC000510192) were deposited at the National Institute of Biological Resources (NIBR) as metabolically inactive cultures.
Cultural and morphological characterization
The strain KNUF-21-66Q1 was cultured and incubated on different media to investigate its cultural and morphological characteristics. The strain was transferred onto PDA, oatmeal agar (OA; Difco, Detroit, MI, USA), malt extract agar (MEA; Difco, Detroit, MI, USA), and cornmeal agar (CMA; Difco, Detroit, MI, USA) and incubated at 25℃ for 10-21 days [3,9]. Different parameters such as colony color, size, shape, and growth were measured and recorded on each different media. The morphological characteristics were observed under a light microscope (BX-50, Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Total genomic DNA from the fungal isolates KNUF-20-NI011 and KNUF-20-NI006 were extracted from the fungal mycelia grown on the PDA plate using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s protocol. For the polymerase chain reaction (PCRmax, Alpha Cycler AC-1, Staffordshire, UK), the ITS1F and ITS4 primer pair was used to amplify the internal transcribed spacer (ITS) regions [16,17], primers NL1 and NL4 were used for partial gene sequences of the 28S rDNA large subunit (LSU) [18], and a fragment of β-tubulin (TUB2) was amplified using the primers T1 and T22 [19]. PCR amplification protocols were performed with slight modification as described [20]. The quality of PCR products was evaluated by electrophoresis on 1.2% agarose gels stained with ethidium bromide. Then PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by SolGent (Daejeon, Korea).
Molecular phylogenetic analyses
The obtained sequences were compared with the reference sequences retrieved from the GenBank database of the National Center for Biotechnology Information (NCBI) (Table 1). Alignments of each gene were performed, the sequences were combined to reveal the position of the isolate, and evolutionary distance matrices for the neighbor-joining algorithm were generated using Kimura’s two-parameter model [17]. Phylogenetic analysis was performed using the MEGA 7.0 program with the bootstrap values based on 1000 replicates [18].
RESULTS AND DISCUSSION
Paraconiothyrium kelleni Santelices, Campos-Quiroz, Carrasco-Fernández, M. Guerra & J.F. Castro, Persoonia 49: 305 (2022) [MB#845530]
Specimen collection: Bibongsan, Jecheon, Chungcheongbuk-do, Korea (37°01ʹ37ʺN, 127°16ʹ45ʺE), isolated from soil.
Morphology of KNUF-21-66Q1
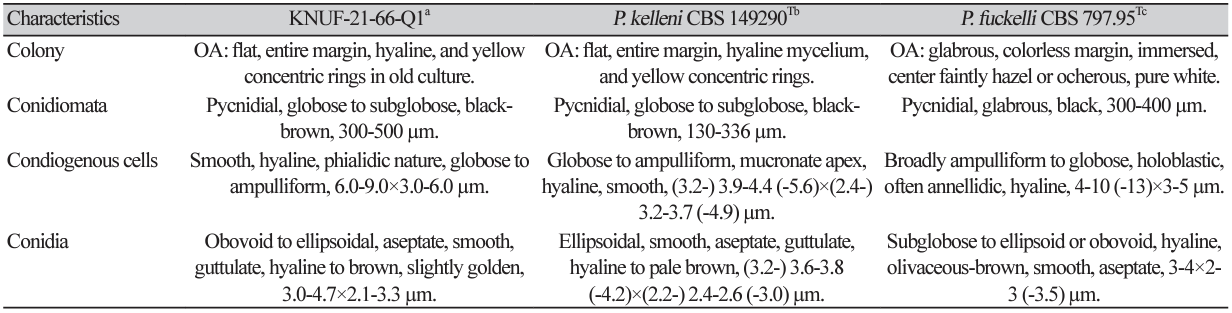
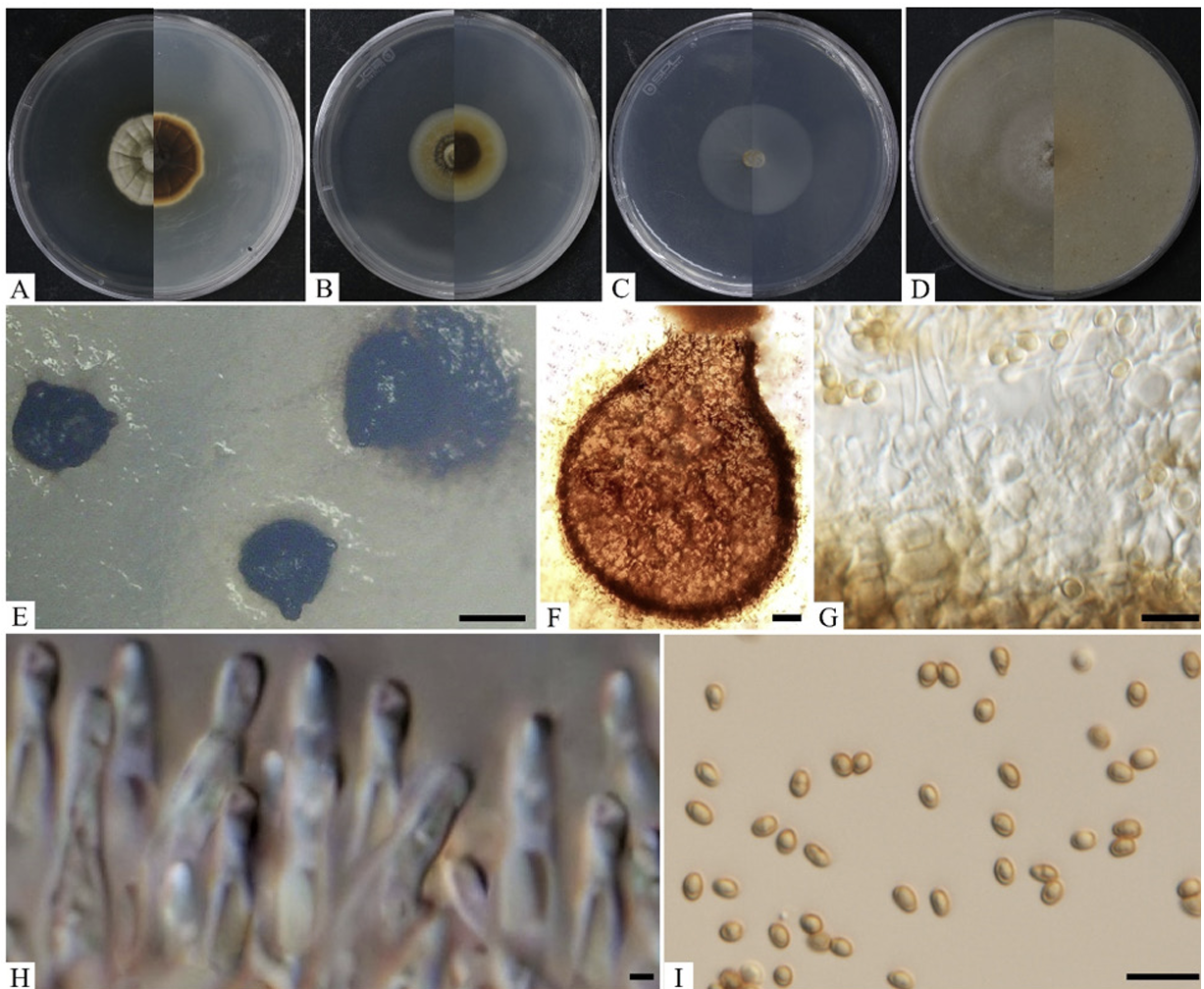
Cultural characteristics: The growth of the colony of strain KNUF-21-66Q1 was moderate on PDA, OA, MEA, and CMA after 10 days of incubation at 25℃ (Fig. 1). Its cultural and morphological characteristics were compared with those of the previously described strain P. kelleni (Table 2). Colonies grown on PDA were 31-33 mm in diameter in 10 days and had an even, pale orange, white margin followed by light to dark brown showing a concentric and radiating pattern. The reverse side showed buff margin and white to gray circular zones toward the center (Fig. 1A). Colonies grown on MEA were 3436 mm in diameter in 10 days showing white, creamy white near the edge and pale yellow to dark brown toward the center. The reverse side showed an umber brown in center surrounded by pale fulvous (Fig. 1B). Similarly, on CMA, the colony reached 50-51 mm in diameter in 10 days and showed a transparent white color, flat mycelium, with the reverse side also showing the same features (Fig. 1C). On OA, the colonies reached 48-49 mm in diameter in 10 days and exhibited flat, entire margin, areas of sparse hyaline mycelium, and yellow concentric rings in old culture. The reverse side showed a colorless margin with white wooly floccose and fluffy aerial mycelium (Fig. 1D).
Morphological characteristics: Conidiomata were asexual morphs that were pycnidial, discrete, mostly submerged in the media, and soft, solitary, or clustered, ostiolate, measuring 300–500 µm in diameter with a shape of globose to subglobose, black-brown in color (Fig. 1E). Pycnidium was observed after 5–6 weeks, showing a thick wall yellowish in color (Figs. 1F and 1G). Conidiogenous cells were smooth, hyaline, and f lat at the base and tapered considerably toward the tip with phialidic nature and globose to ampulliform, and the size ranged from 6.0-9.0×3.0-6.0 µm (n=15). Conidiophores were not observed separately and reduced to conidiogenous cells (Fig. 1H). Conidia were hyaline and later turned pale brown, slightly golden when matured, obovoid to slightly ellipsoidal, aseptate, smooth, and guttulate, although non-guttulate conidia were also observed. The conidia size was 3.0-4.7×2.1-3.3 µm (n=100) with an average length-towidth (L/W) ratio of 1.6 (Fig. 1I).
Fig. 1
Cultural and morphological characteristics of KNUF-21-66Q1. Colonies on potato dextrose agar (PDA) (A), malt extract agar (MEA) (B), cornmeal agar (CMA) (C), and oatmeal agar (OA) (D), front and reverse, respectively, after 10 days at 25℃. Conidiomata after 21 days at 25℃ from OA (E), pycnidium (F), pycnidial wall (G), conidiogenous cells (H), and conidia (I). Scale bars: E=200 μm, F=20 μm; G-I=10 μm.
Molecular phylogeny of KNUF-21-66Q1
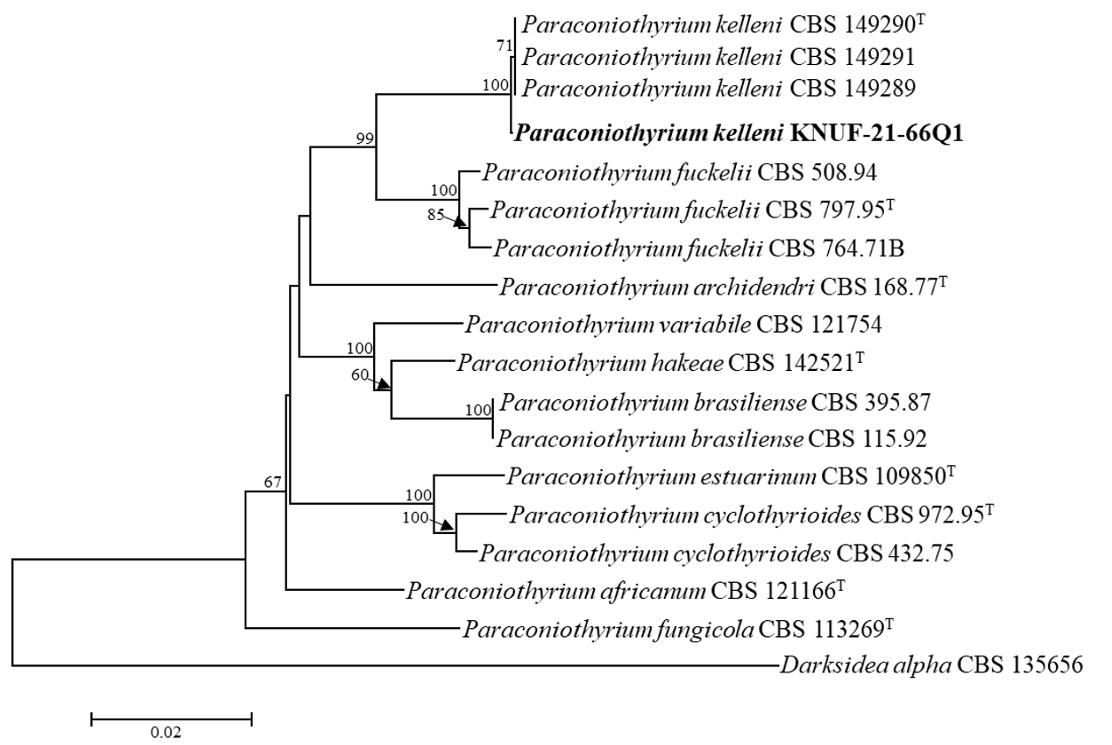
The NCBI database from the BLAST search showed that the ITS (569 bp) regions sequences of the strain KNUF-21-66Q1 exhibited a high similarity of 99.8% with the different strains of P. kelleni (CBS 149289, CBS 149290T, and CBS 149291), followed by maximum similarities of 95.0-95.8% with P. variabile (JAC12363, 18EPLE022, CBS 121163, and CBS 680.83). The LSU sequences (815 bp) of the strain shared 100% similarity with the strains of P. kelleni (CBS 149289, CBS 149290T, and CBS 149291), 99.8-99.9% similarities with several strains of P. fuckelii (T235I1, CBS 797.95, CBS 764.71B, and CBS 653.85), and P. rosae MFLU 15-1115. TUB2 (620 bp) exhibited the highest similarity of 99.3-99.8% with the strains of P. kelleni (CBS 149289, CBS 149290T, and CBS 149291), followed by maximum similarities of 89.5-91.7% with the strains of P. fuckelii (CVG482, JZB320004, CBS 653.85, and CBS 797.95). Concatenated datasets of ITS, LSU, and TUB2 sequences were used to determine the molecular relationship between the isolated Korean strain and closely related Paraconiothyrium species (Fig. 2). The tree topology of the neighbor-joining method in the phylogenetic analysis confirmed that the strain KNUF21-66Q1 was closely clustered with the previously identified P. kelleni. Hence, KNUF-21-66Q1 was identified as P. kelleni, which was isolated from soil in Korea.
Fig. 2
Neighbor-joining phylogenetic analysis of KNUF-21-66Q1 based on combined sequence data of β-tubulin (TUB2), 28S rDNA of large subunit (LSU), and internal transcribed spacer (ITS) regions showing the phylogenetic position among the closest species of Paraconiothyrium. The strain isolated in this study is in bold, and the bootstrap values ( >50%) are based on 1000 replicates. Darksidea alpha CBS 135656 was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
Paraconiothyrium species are found to be associated with wilting of grapes, trunk rots of vines, pear, and apple trees, and cane blight of raspberry [19-21]. Apart from plant pathogens, species such as P. cyclothyrioides are associated with the causative agent of human cutaneous phaeohyphomycosis [22]. Three unreported Paraconiothyrium species have already been recorded from Korea, viz., P. brasiliense from Chinese maple leaf [23], P. archidendri from plant litter in freshwater, and P. fuckelii from the wild plant Rubus oldhamii [24,25]. However, some of the Paraconiothyrium species are rationally utilized for biocontrol, bioremediaton, and antibiotic production. Pathogens such as Phytophthora spp. and Colletotrichum spp. have been found to exert an antagonistic effect with P. brasiliense [26]. Moreover, various bioactive compounds were isolated from Paraconiothyrium species, such as the antitumour drug “Paclitaxel,” also known as taxol, produced by endophytic fungi P. brasiliense and P. variabile and used to treat various cancers [7,27,28]. In total, 150 secondary metabolites have been isolated from Paraconiothyrium species, including novel diterpenes, sesquiterpenes, polyketides, and aromatic compounds [5].
In conclusion, further research is required to investigate the etiology, pathogenicity of P. kelleni and the sources of secondary metabolites based on the ecological and environmental conditions in Korea. To the best of our knowledge, this is the first record of P. kelleni in Korea.