INTRODUCTION
Airborne environments harbor a diverse array of biological aerosols, including bacteria, fungi, pollen, and tissues [1]. Fungal spores, particularly in outdoor and indoor air, constitute a significant focus of research in the field of aerobiology [2]. Previous studies focus on the sources and dispersal patterns of airborne fungi, temporal variations in fungal species concentrations, and their relationships with other sources. The composition of fungal communities in the air varies considerably across continents and oceans [3]. In Korea, a nationwide study identified Cladosporium as the most abundant airborne fungus, followed by Alternaria and Irpex [4].
Penicillium fungi are ubiquitous, with species found in diverse environments, including indoor and outdoor air, soil, marine habitats, and foods [5]. Among the 483 currently documented species, Penicillium is a broad genus classified into 2 subgenera and 32 sections by Houbraken et al. [6]. The systematics of Penicillium species were determined through the analysis of morphological characteristics, such as colony features, and the number and shape of conidiophore branches. Molecular phylogeny relied on DNA regions, primarily the internal transcribed spacer (ITS), β-tubulin (BenA), calmodulin (CaM), and RNA polymerase II second-largest subunit (RPB2) [7].
In Korea, numerous species of Penicillium have been documented over the years, and a comprehensive list was compiled by Kim et al. [8]. Recently, 15 previously unrecorded species were identified [912]. However, the number of reported Penicillium species in Korea remains lower than the global count. Considering the industrial significance of Penicillium species, particularly in antibiotic and food production, as well as their potential pathogenicity to crops and mycotoxin production [13], ongoing studies on Penicillium species in Korea are crucial.
In this study, we report the occurrence of P. mexicanum, which has not previously been reported in Korea. We isolated this strain from air samples and conducted morphological identification in conjunction with phylogenetic analyses utilizing the ITS, BenA, and CaM regions.
MATERIAL AND METHODS
Sample collection and fungal isolation
In 2020, air samples were collected from Anmyeon-eub, Taean-gun, Chungcheongnam-do, Korea (36°29'52.1"N 126°21'11.1"E). Airborne fungi were captured by exposing petri dishes containing potato dextrose agar (PDA) medium. The plates were subsequently incubated at 25℃ until the appearance of single colonies. A single colony was then transferred to a fresh PDA plate and further incubated at 25℃. Strain Y20P-5 was chosen based on cultural, morphological, and phylogenetic analyses, and it was subsequently deposited at the Mycology Laboratory, Korea National University of Education.
Cultural and morphological characteristics
The isolated strain was cultured and incubated on various growth media, including PDA, malt extract agar (MEA), Czapek yeast autolysate agar (CYA), dichloran 18 % glycerol agar (DG18), and oatmeal agar (OA), to investigate its cultural and morphological characteristics. The strain was incubated at 25℃ for 7 days. After incubation, various colony characteristics, such as color, size, shape, and growth pattern, were measured and recorded for each medium. Morphological features of the fungi were observed under a light microscope using the slide culture method on PDA, MEA, and water agar (WA). The dimensions of the conidia and the length and diameter of the conidiophores were measured using an eXcope.
Genomic DNA extraction, PCR, and sequencing
Genomic DNA was extracted from the mycelia of isolated strain Y20P-5 using the HiGene Genomic DNA Prep Kit (BioFACT, Daejeon, Korea) following the manufacturer’s instructions. The ITS region was amplified using the ITS1F/ITS4 primers [14,15], the BenA region was amplified using the bt2a/bt2b primers [16], and the CaM region was amplified using the cmd5/cmd6 primers [17]. Amplified DNA was sequenced by SolGent Co., Ltd. (Daejeon, Korea). The DNA sequences were submitted to GenBank, National Center for Biotechnology Information (NCBI).
Molecular phylogenetic analyses
The obtained nucleotide sequences were analyzed for species identification using the Basic Local Alignment Search Tool (BLAST), NCBI. The sequences of the ITS, BenA, and CaM regions were combined, and phylogenetic trees were constructed using the maximum likelihood method with an adjusted Kimura-2-parameter model and 1,000 replication bootstraps in MEGA7 software [18]. Reference sequences from the GenBank database of NCBI (Table 1) were utilized for the analysis.
RESULTS AND DISCUSSION
Penicillium mexicanum Visagie, Seifert & Samson, Studies in Mycology 78: 125 (2014) [MB#809185]
Specimen examined: Anmyeon-eub, Taean-gun, Chungcheongnam-do, Korea (36°29'52.1"N 126° 21'11.1"E), isolated from air, strain Y20P-5, GenBank accession numbers OQ048471 (ITS), OQ130423 (BenA), OQ134945 (CaM).
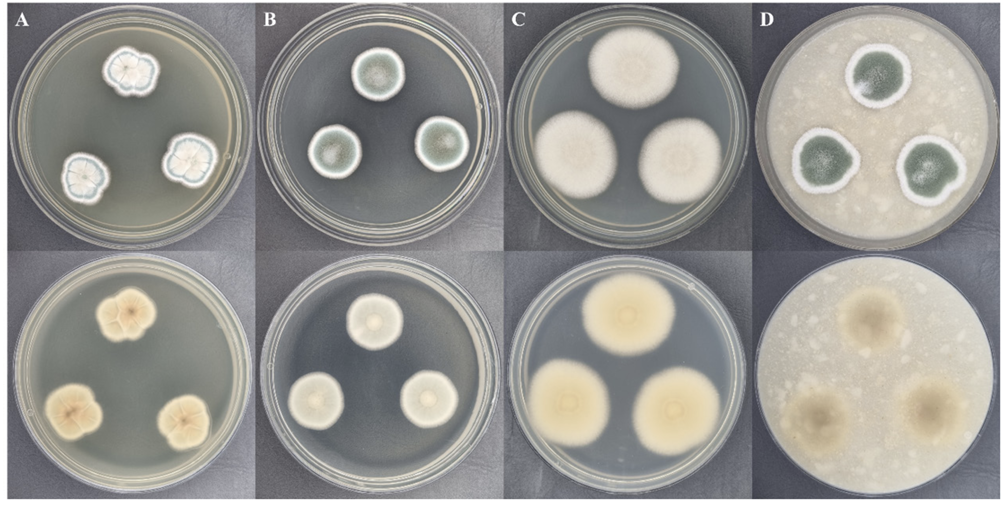
Cultural characteristics: Penicillium mexicanum Y20P-5 was cultured on CYA, DG18, MEA, and OA media at 25℃ in the dark for seven days. Colony diameters were observed to be 17.8-20.7 mm on CYA, 21.3-22.3 mm on DG18, 31.8-33.1 mm on MEA, and 26.1-27.3 mm on OA (Fig. 1). The colony on CYA exhibited an irregular shape with undulate margins, a filamentous appearance, ivory color at the center, teal color at the conidial ring, white color at the margin, wrinkled and powdery texture, and a light brown reverse (Fig. 1A). The colony on DG18 was circular with even margins, slightly filamentous, greyish green in color, white at the edge, little cottony texture at the center, powdery texture, and an ivory reverse (Fig. 1B). The colony on MEA was circular and lightly ellipsoidal, with even margins, slightly filamentous, ivory color, powdery texture, and a light brown reverse (Fig. 1C). The colony on OA exhibited a circular or irregular shape with slightly filamentous margins, greyish green color at the center, white color at the edge, little cottony texture, and powdery appearance (Fig. 1D).
Fig. 1
Colonies of Penicillium mexicanum Y20P-5 cultured at 25℃ in dark for 7 days. (A) Czapek yeast autolysate agar (CYA); (B) Dichloran 18% glycerol agar (DG18); (C) Malt extract agar (MEA); (D) Oatmeal agar (OA). Top is front side; bottom is reverse side.
Morphological Characteristics: The conidiophores were terverticillate (Fig. 2, Table 2). The stipes ranged from 65.3 to 127.6 μm in length and 2.7 to 4.0 μm in width, with 2-3 branches per stipe. The dimensions of the branches were 5.2-17.8 μm in length and 2.8-4.1 μm in width. The metulae measured 5.9-14.3 μm in length and 2.3-4.0 μm in width (average 8.8±1.8 μm in length and 2.9±0.5 μm in width). The phialides were flask-shaped and measured 4.4-10.4 μm in length and 2.0-3.7 μm in width (average 6.4±1.2 μm in length and 2.6±0.5 μm in width, n=45). The conidia were subglobose or ellipsoidal, with dimensions of 2.7-4.7 μm in length and 2.5-3.7 μm in width (average 3.2±0.3 μm in length and 2.8±0.3 μm in width, n=45). On CYA, DG18, and OA media, the conidia exhibited a teal or grey-green color.
Molecular phylogeny: Molecular phylogenetic analysis revealed a high sequence similarity between the strain and P. mexicanum. The ITS region exhibited a 99.8% match to the type strain of P. mexicanum CBS 138227 (NR_137878), the BenA region showed a 98.5% match to the type strain of P. mexicanum DTO 270F1 (KJ775178), and the CaM region exhibited a 95.4% match to the type strain of P. mexicanum DTO 270F1 (KJ775412). In the phylogenetic tree constructed using the maximum likelihood method (Fig. 3), the Y20P-5 sequence formed a monophyletic group with P. mexicanum DTO 270F1.
Penicillium mexicanum was initially documented in 2014 and was reported to have been isolated from house dust in Mexico [19]. Its conidiophores were treverticillate, with 2-3 branches diverging from a single stipe. The average size of the conidia, over 3 μm, aligned with the major morphological differences described in the original description of P. atramentosum. Therefore, based on both morphological characteristics and molecular phylogenetic analysis, the isolated fungus was confirmed as P. mexicanum. This study represents the first report of P. mexicanum in Korea.
Fig. 2
Conidia and conidiophores of Penicillium mexicanum Y20P-5. (A) Conidia. (B-D) Conidiophores. Scale bars: A, B=10 μm; C, D=50 μm.
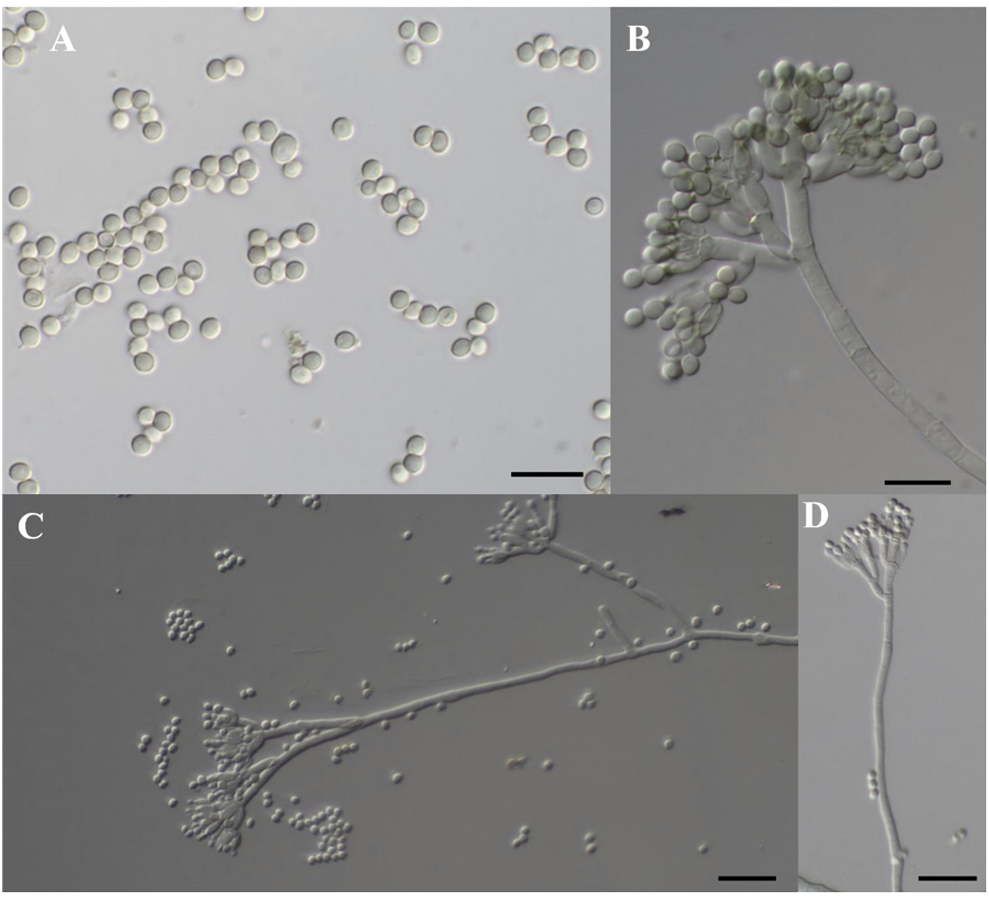
Table 2
Comparison of morphological characteristics of Penicillium mexicanum strains Y20P-5 and CBS 138227.
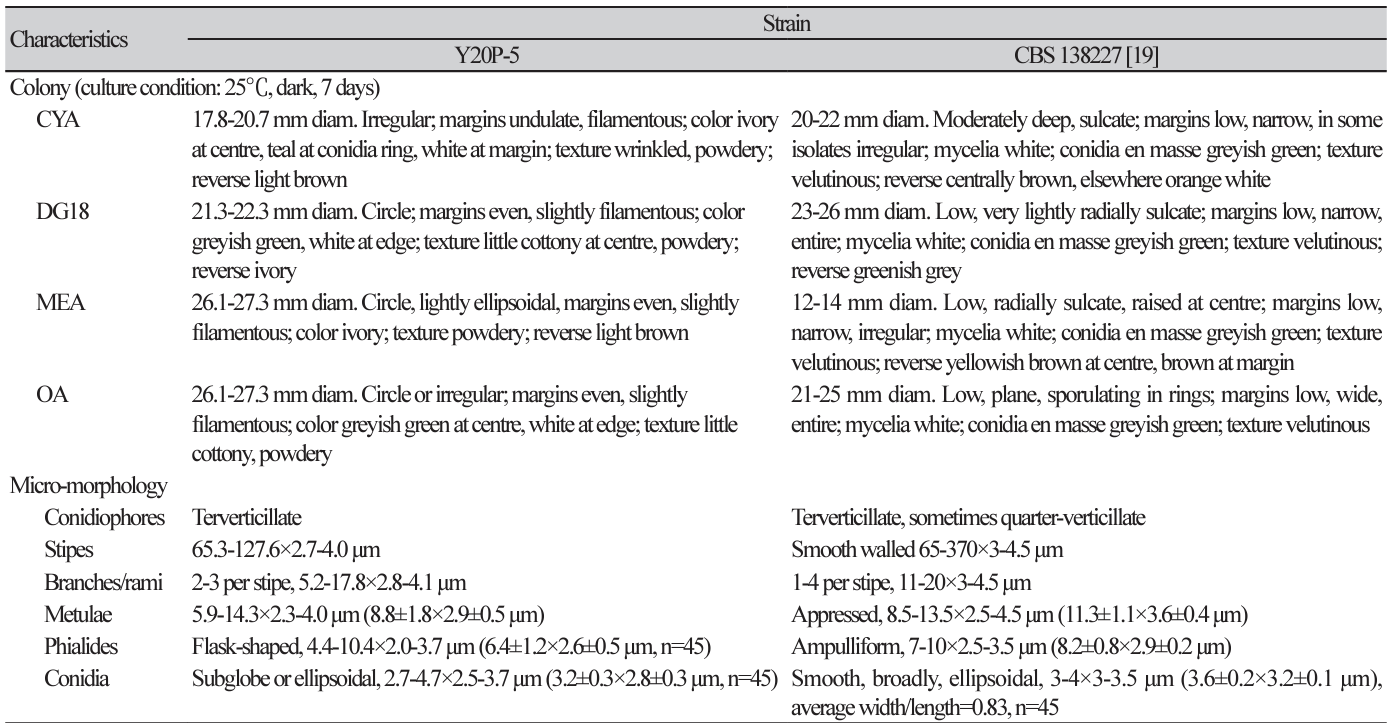
Fig. 3
Maximum likelihood phylogenetic analysis of combined internal transcribed spacer (ITS), β-tubulin (BenA), and calmodulin (CaM) sequences. Talaromyces piceae was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in bold.