Fatoua villosa (Thunb.) Nakai, commonly known as mulberryweed or hairy crabweed, belongs to the family Moraceae. It is native to Eastern Asia and was introduced to Louisiana, USA, in the 1950s. Given its ability to reproduce and spread rapidly, it has been naturalized throughout much of the eastern United States and is found from Texas to Florida and north of Michigan and Delaware [1-3]. This species was recently reported in Australia (Queensland) and Europe (France) [https://gd.eppo.int/taxon/FTOVI].
To eradicate this troublesome weed, multiple attempts have been made using chemicals but not effective biological tools [4,5]. Some phytopathogenic fungi, such as Pseudocercospora fatouae Goh & W.H. Hsieh. in China and Taiwan, Podosphaera pseudofusca (U. Braun) U. Braun & S. Takam. in China and Japan, and Podosphaera xanthii (Castagne) U. Braun & Shishkoff in Korea, have been identified on this weed [6,7]. Given that P. fatouae is a culturable fungus, it can be considered a potential biocontrol agent. However, its distribution and biology are poorly understood. The purpose of this study was the morphological and molecular characterization of the fungus on F. villosa in Korea.
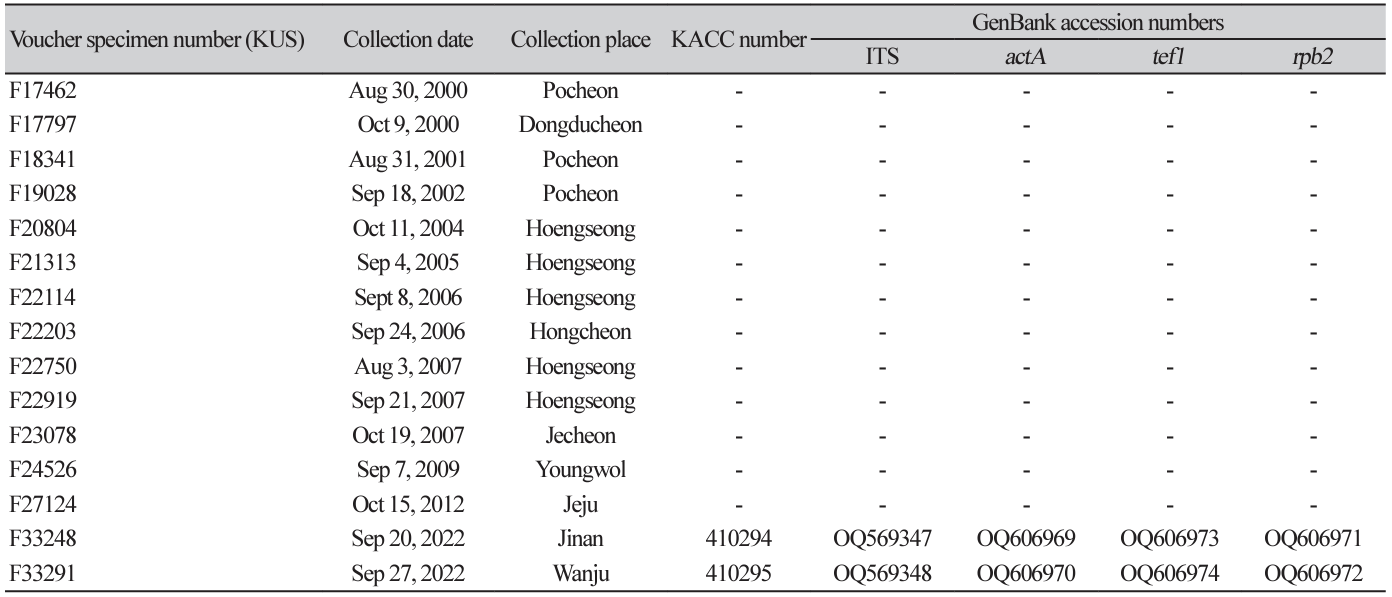
During our field forays in Korea, fifteen samples of fungus-infected F. villosa were collected and preserved in the Korea University herbarium (KUS) (Table 1). Preliminary microscopy confirmed the fungus on these samples as P. fatouae based on the characteristic growth pattern on the lower leaf surface and the morphology of conidiophores and long conidia. Detailed morphological characterization and molecular analysis were performed using two samples collected in 2022.
For morphological investigations, a small amount of fungus was scraped from fresh samples, mounted in a drop of sterile water, and examined under an optical microscope (Equipped with a KCS-3.1C Imaging System, Carl Zeiss AX10, Göttingen, Germany). A minimum of 30 measurements were performed for each diagnostic structure. To obtain a pure conidial culture, conidia were collected from lesions using a sterile scalpel and placed in Eppendorf tubes containing sterile water. The conidial suspension was streaked onto 2% water agar (WA, Junsei, Tokyo, Japan) plates supplemented with 100 mg/L of streptomycin sulfate and incubated at 25℃. After two days, colonies were transferred onto potato dextrose agar (PDA, Difco, France) plates. The isolates were obtained from KUS-F33248 and F33291 and submitted to the Korean Agricultural Culture Collection of the Rural Development Administration.
Symptoms initially appeared as pale greenish or yellowish foliar spots on the upper leaf surface, corresponding to dark-gray fuliginous growth on the lower surface. With disease progression, the leaf lesions became vein-limited and angular (Fig. 1A). Ultimately, by coalescing the leaf lesions, more than half of the leaf surface was covered with chocolate-colored fungal growth (Fig. 1B). Stromata were nearly absent or contained only aggregates of several swollen cells in the stomatal openings (Figs. 1C and D). Conidiophores were emerging from stomatal openings, 2-10 in loosely divergent fascicles, olivaceous brown, darker than conidia, 0-3 times branching, tortuous and geniculate, sinuous, 3-6-septate, 30-70 µm long, 3-5 µm wide, and conidial scars inconspicuous (Figs. 1E and F). Conidia were solitary, filiform to cylindrical, pale olivaceous, substraight or slightly curved, 4-10-septate, apex subobtuse, base subtruncate, 50-110 µm long, 3.5-5.0 µm wide, and hilum unthickened and inconspicuous (Figs. 1G and H). Two-weekold fungal colonies grown on PDA at 25℃ measured 23-25 mm in diameter and were olivaceous gray to khaki with smooth undulate margins (Fig. 1I). The reverse culture was green-black. These characteristics are similar to those of P. fatouae Goh & W. H. Hsieh [8].
Fig. 1
Pseudocercospora fatouae hypophyllous leaf mold on Fatoua villosa. (A) Early symptoms of angular leaf mold on the lower leaf surface. (B) Leaf symptoms in the later stage of disease development. Note the leaf mold on the lower leaf surface (LM), yellow discoloration on the upper leaf surface (YD), and necrosis of the affected leaves (NC). (C) A close-up view of the lower leaf surface with numerous conidiophores. (D, E) Conidiophores emerging from the stomatal openings. (F) Conidiophore showing branched character. (G) Conidiophores with small stromata and a young conidium on the tip (arrow). (H) Conidia. (I) Three-week-old colonies of P. fatouae growing on potato dextrose agar at 25℃.

For molecular analysis, genomic DNA was extracted from 2-week-old cultures grown on PDA at 25℃ using MaglistoTM 5M kits (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol. The internal transcribed spacer (ITS) region, including protein coding genes such as, actin (actA), translation elongation factor 1-alpha (tef1), and DNA-directed RNA polymerase II second largest subunit (rpb2), was amplified and sequenced using primer pairs V9G/ITS4, ACT-512F/ACT-783R and EF1-728F/EF1-986R through the polymerase chain reaction (PCR) steps described previously by Nakashima et al. [9]. Amplification of rpb2 gene was not successful with RPB2-5f2/fRPB2-7cR or fRPB2-5F/fRPB2-7cR. As a result, we developed the following primers for amplification of this gene: forward P-RPB2-F (5ʹ-ACC ACT CCC ATC GGT CGA GA-3ʹ) and reverse P-RPB2-R (5ʹ-GGT GTC ATG CTG ATC ATA GC-3ʹ); the PCR procedure was as follows: initial denaturation (94℃ for 2 min), five amplification cycles at 94℃ for 45 s, 60℃ for 45 s, and 72℃ for 45 s, followed by 30 amplification cycles at 95℃ for 45 s, 54℃ for 45 s, and 72℃ for 45 s, and a final extension at 72℃ for 5 min). The PCR products were sequenced in both directions using the same primers by a commercial sequencing company (Bioneer, Daejeon, Korea). Forward and reverse sequences were assembled, and the consensus sequences were deposited in GenBank (Table).
The alignment for each gene was prepared separately in MEGA 11 [10] and then combined into one multigene dataset of ITS+act+tef1+rpb2 using SequenceMatrix 1.8 software [11]. Trochophora simplex (CBS 214744) and Pallidocercospora heimioides (CBS 111190) were selected as outgroups. Maximum parsimony analysis was conducted in PAUP* 4.0a using a heuristic search option [12]. The robustness of the tree was evaluated using bootstrap analysis with 1,000 replicates.
The resulting sequences obtained from the two samples were almost identical; only a one-bp difference was found in actA. A BLASTn search was performed for newly obtained sequences to compare their similarity with other reference sequences in the GenBank. The findings revealed 100% identity to Pseudocercospora crocea (MH863878, GU384502) for the ITS and tef1 genes, whereas for actA, 99.5% similarity to P. balsaminae (GU320367) and P. farfugii (LC599423) and for rpb2, 99.84% similarity to P. farfugii (LC599603) was observed.
The final alignment consisted of 57 sequences and 1,846 characters, of which 187 (10.13%) were variable and parsimony uninformative, and 605 (32.77%) were informative for parsimony analysis. As demonstrated in the resulting tree, the sequences of P. fatouae found on F. villosa were placed in a distinct clade with those of P. crocea; this was supported by a 100% bootstrap value (Fig. 2). Given the lack of rpb2 sequences for P. crocea (CBS126004) in GenBank, only the sequences of ITS (GU269792), actA (GU320493), and tef1 (GU320493) obtained from that isolate were included in the current analysis. However, P. crocea described on Pilea hamaoi (Urticaceae) in 2013 [13] differs from the current species by having amphigenous caespituli (vs. hypogenous caespituli), a well-developed stroma of 40-100 µm diam. (vs. lacking or only an aggregate of several swollen cells), and shorter conidiophores, 17-24 µm long (vs. 30-70 µm long). As a result, P. crocea is not close to, or synonymous with, P. fatouae.
Fig. 2
A phylogenetic tree of Pseudocercospora fatouae was constructed using the maximum parsimony method based on a combined multigene dataset of ITS+actA+tef1+rpb2 of 59 sequence samples. The isolates obtained in this study are shown in boldface. Bootstrap values (>70%) were provided on the relevant branches. Tree scores, such as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC), are presented in the box given on the left side of the Figure. ITS, internal transcribed spacer; actA, actin; tef1, translation elongation factor 1-alpha; rpb2, DNA-directed RNA polymerase II second-largest subunit.

P. fatouae was first described by Hsieh & Goh [8] as a new species in Taiwan in 1990. Previously, Cercospora fatouae (originally known as C. fatuae) was described as a new species in Japan in 1904 [14]. The morphological description of C. fatouae is extremely close to that of P. fatouae in having hypogenous fructification with dark fuliginous fungal growth, branched conidiophores, and long conidia (90-120×3-4 µm) in the same genus, Fatoua spinosa var. subcordata, indicating that these two species are synonymous. Therefore, we propose the correct name for this fungus as Pseudocercospora fatouae (Henn.) Goh & W. H. Hsieh.
Alien-invasive plants are of great concern worldwide. Mulberryweed (F. villosa) is an Asian plant that is invasive in North America and, recently, in France. Understanding the diversity of phytopathogenic fungi on mulberryweed in its indigenous regions is critical for understanding this weed and developing effective control measures. Leaf mold (P. fatouae) on mulberry weeds has been reported in China and Taiwan [7]. To the best of our knowledge, this is the first report of P. fatouae infection in F. villosa in Korea. This study contributes to the understanding of the biology of P. fatouae by providing morphological characteristics and molecular phylogenetic data, as well as depositing two monoconidial isolates from Korea in the culture collection.