INTRODUCTION
Zygomycetous fungi, such as Mortierellomycota and Mucoromycota, are classified based on their reproductive characteristics. They produce zygospores for sexual reproduction, and conduct asexual reproduction via sporangia [1]. In yesteryears, the two phyla were grouped in a same phylum, Zygomycota; however, recently, they have been classified as two separate phyla [2]. Several species belonging to Mortierellomycota and Mucoromycota have been isolated from the soil, plant roots, decaying plant materials, endophytic plant litter, insect guts, mosses, animal dung, and freshwater environments [3,4].
Fungal species belonging to Mortierellomycota generally possess anastomosing hyphae and thalli with dichotomously branching multispored spherical sporangia [2]. They are found worldwide in various habitats including aquatic and terrestrial environments [2]. As stated in Mycobank, 21 genera have been reported as of 2022 [5].
The genus Dissophora of Mortierellaceae contained three species as of 2021 [6]. Using low-coverage genome sequences, this genus forms a clade distinct from Mortierella species [3]. Dissophora fungi are known to be found in forest litter and soil, unlike common Mortierella species [3]. Fungal species of this genus have fertile, septate aerial stolons that are differentiated from vegetative hyphae [3].
In the genus Linnemannia of family Mortierellacae, 26 species have been reported as of 2022 [7]. Numerous fungal species of this genus have been isolated from soils, and are associated with the plant rhizosphere or litter [3]. This genus is classified according to the sporangia characteristics and sporangiospore morphology [3].
In the genus Mortierella of family Mortierellacae, 173 species were reported in 2023 [5]. Many Mortierella species grow well at cooler temperatures and few species have been reported to be mycoparasites [3]. Various Mortierella species have been found in a wide range of habitats, including soil, water, living plant roots, and plant debris [3]. In South Korea, four Mortierella species have been isolated from freshwater sources [8,9].
Several fungal species belonging to Mucoromycota have been reported to be plant-associated fungi (e.g., plant symbionts, plant debris decomposers, or pathogens) [10]. They generally produce globose, smooth, or ornamented zygospores and exhibit rapid mycelial growth [10].
The genus Umbelopsis belongs to the family Umbelopsidaceae, order Umbelosidales, class Umbelopsidomycetes, and phylum Mucoromycota [2]. To date, 22 species of this genus have been reported [11]. Several species have been isolated from the soil, plant debris, roots, and human skin [11,12]. This genus is classified according to characteristics such as the morphology of the sporangia, sporangiospores, and colonies [12].
This is the first report of four fungal species (namely Dissophora globulifera, Linnemannia exigua, Mortierella rishikesha, and Umbelopsis autotrophica) found in South Korea from environmental samples such as freshwater, plant litter, and sediment in streams. The molecular, phylogenetic, and morphological characteristics of these species were also investigated.
MATERIALS AND METHODS
Isolation of fungal strains
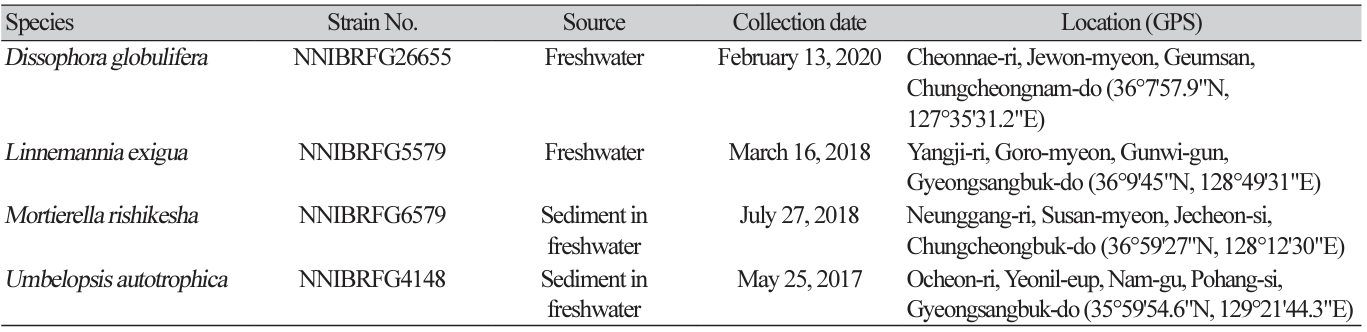
Fungal strains were isolated from the freshwater environmental samples (filtered water and stream and river sediments) collected in South Korea. The collection information for all the strains identified in this study is listed in Table 1. A dilution method was used to isolate fungal strains from the sediment. The diluted suspensions (200 μL) of sediment or soil with distilled water (1:100 and 1:1,000) were spread on potato dextrose agar (PDA; 3.9% PDA powder [w/v]; Difco, Sparks, MD, USA) containing 100 ppm of streptomycin, and fungal strains were isolated in pure form after incubation for 4-5 days at 25℃ by repeating this step. To isolate fungal strains from freshwater, 50 mL of freshwater was filtered through a membrane filter. Membrane filter was attached on water agar (WA; 1% agar powder; Duksan Pure Chemicals Co., Ltd., Ansan, Korea) with 100 ppm of streptomycin and incubated at 20℃ overnight. After removing membrane, hyphal tips and germinated conidia were observed under a microscope, transferred onto a 24-well plate containing V8 agar (V8A; 8% V8 juice [v/v] and 1.5% agar [w/v], adjusted to pH 6.0 using 10 N NaOH), and incubated for 7 days at 25℃ in the dark. All strains identified in this study were grown on malt extract agar (MEA; 2% malt extract [w/v] and 2% agar [w/v]), oatmeal agar (OA; 7.25% OA powder [w/v]; Difco), corn meal dextrose agar (CMDA; 2% cornmeal [w/v], 2% glucose [w/v], and 2% agar), and yeast extract peptone dextrose agar (YPDA; Duchefa Biochemie, Haarlem, Netherlands).
DNA extraction, polymerase chain reaction (PCR), and DNA sequencing
Fungal genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Düren, Germany). For the molecular identification of fungi, PCR amplification was performed for the internal transcribed spacer (ITS) rDNA region using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [13], and the large subunit of rDNA (LSU) was identified using primers LR0R (5′-ACCCGCTGAACTTAAGC-3’) and LR7 (5′-TACTACCACCAAGATCT-3′) [14]. Amplicons were sequenced by a DNA sequencing service (Macrogen Inc., Seoul, Korea) using the same primers as those used for amplification. A homology search of the DNA sequences was conducted using BLAST algorithms from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm. nih.gov).
Phylogenetic analysis
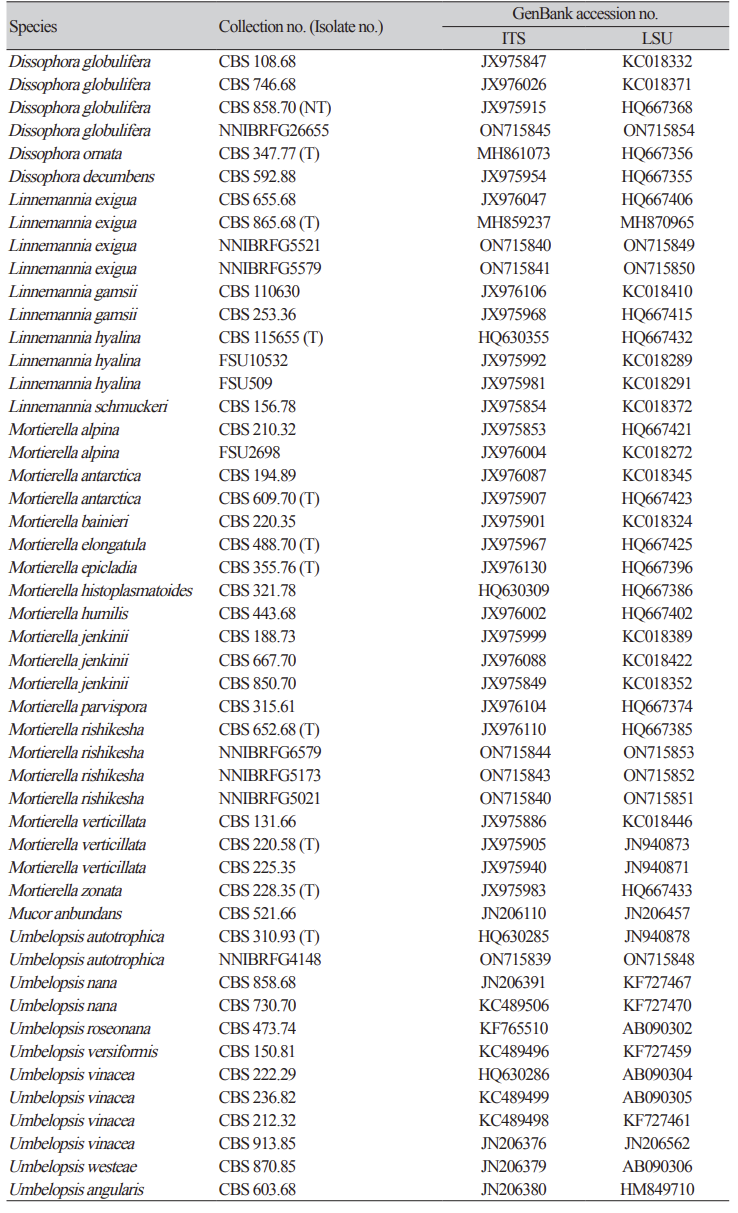
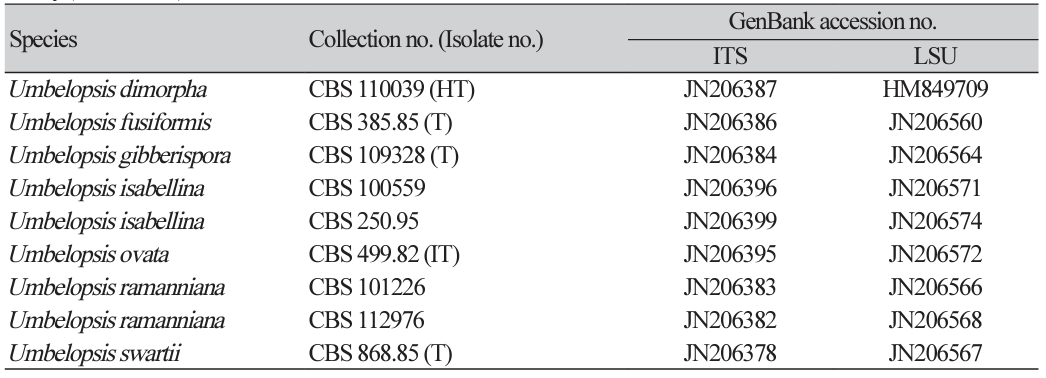
Previously published reference sequences [3-4,7,9-12,15] were obtained from the NCBI database (Table 2). The sequences were edited using DNAStar (version 5.05, DNAStar Inc., Madison, WI, USA). The accession numbers of the sequences used in this study are listed in Table 2. Phylogenetic trees were constructed using maximum likelihood (ML) analysis, which was performed using the default settings of MEGA 7.1 [16], except for the replacement with Tamura-Nei model. Bootstrapping analysis of 1,000 replicates was performed to test the robustness of each grouping.
Morphological analysis
The microstructures of the fungal species were observed under an Eclipse Ni light microscope (Nikon, Tokyo, Japan) equipped with a Ds-Ri2 digital camera (Nikon). At least 50 individuals were examined to observe and measure each structure. For scanning electron microscopy, we followed the protocol described by Alves et al. [17]. Photographs were captured using a scanning electron microscope (SEM; SU8220, Hitachi, Tokyo, Japan).
RESULTS AND DISCUSSION
Phylogenetic analysis
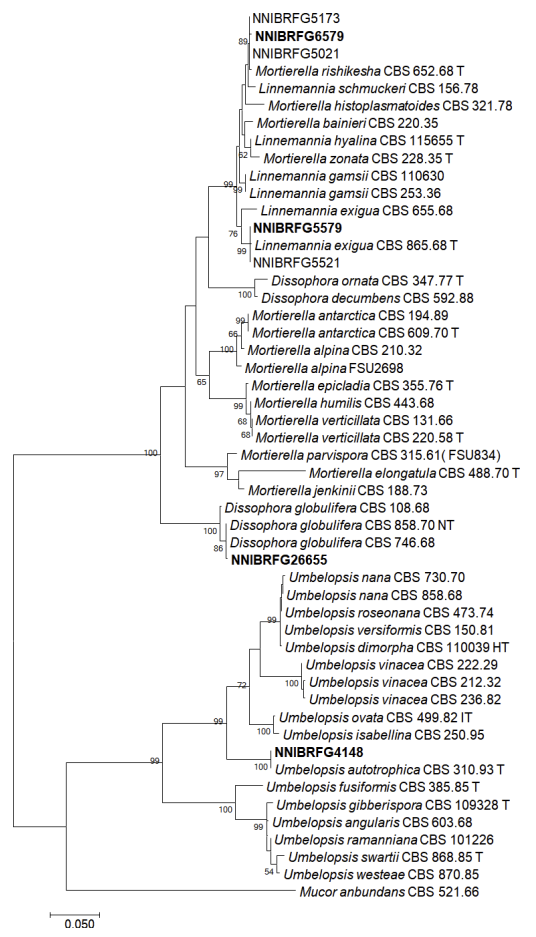
Phylogenetic analysis, using a combination of ITS and an LSU, was performed to identify fungal strains and infer their phylogenetic relationships with other similar species. As shown in Fig. 1, the strain NNIBRFG6579 formed a clade with M. rishikesha strain CBS 652.68. The sequences of NNIBRFG6579 showed high similarity–100% (ITS), 99.69% (LSU), and 100% (EF1)–to those of M. rishikesha. NNIBRFG5579 formed a clade with L. exigua strain CBS 965.68. The sequences of NNIBRFG5579 showed high similarity–99.69% (ITS) and 99.42% (LSU)–to those of L. exigua. NNIBRFG26655 formed a clade with CBS 858.70 of D. globulifera. The sequences of NNIBRFG26655 showed high similarity–99.52% (ITS) and 99.90% (LSU)–to those of D. globulifera. NNIBRFG4148 formed a clade with the CBS 310.63 of U. autotrophica. The sequences of NNIBRFG4148 showed high similarity–100% (ITS) and 100% (LSU)–to those of U. autotrophica.
Fig. 1
Phylogenetic tree of Dissophora globulifera, Linnemannia exigua, Mortierella rishikesha, Umbelopsis autotrophica and their related species, based on maximum-likelihood analysis of the combination of internal transcribed region sequences (ITS) and large subunit of ribosomal DNA (LSU) sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. Mucor abundans is outgroup. Bold type font indicated the strains used in this study. T, type material.
Three fungal species of Mortierellomycetes and one species of Umbelopsidomycetes were discovered from freshwater environment and identified through molecular identification.
TAXONOMY
Dissophora globulifera (O. Rostrup) Vandepol & Bonito, Fungal Diversity 104: 279 (2020) [MB#833727] (Fig. 2A, 3A-D)
Description: Colonies grew fast at 25 ℃ and reached 55, 58, 42, 56, 64, and 60 mm on CMDA, MEA, OA, PDA, V8A, and YPDA, respectively, 7 d after inoculation. The colony color was hyaline with a smooth aerial mycelial surface on CMDA and MEA, hyaline-to-creamy white with short aerial mycelia on OA, translucent white with fluffy aerial mycelia showing a concentric pattern on PDA, translucent white with cottony aerial mycelia showing a concentric pattern on V8A, and translucent white with fluffy aerial mycelia on YPDA. Sporangia were hyaline and 19.0-35.3 µm diameter (x=26.8±5.34 µm, n=10). Sporangiospore were hyaline, globose, echinulate, and 6.8-9.0 µm in diameter (x=7.8±0.55 µm, n=50). Chlamydospores were round-to-oval, thick-walled, and 9.3-12.9 µm in diameter (x=11.1±1.45 µm, n=10).
Habitat: Filtered freshwater from a stream
Specimen examined: Cheonnae-ri, Jewon-myeon, Geumsan, Chungcheongnam-do, Republic of Korea; 13 Feb, 2020, NNIBRFG26655, Nakdonggang National Institute of Biological Resources (collected by Jaeduk Goh).
Note: D. globulifera was first described as isolates from soil or organic debris in soil [18]. The type specimen MBT#8101(neotype) was isolated from the decaying root of Dactylis glomerata [3]. Other strains reported as D. globulifera have been isolated from agricultural soil in Netherlands, Schweden, England, and Austria [3]. In this study, NNIBRFG26655 was isolated from filtered freshwater streams.
Linnemannia exigua (Linnem.) Vandepol & Bonito, Fungal Diversity 104: 283 (2020) [MB#835752] (Fig. 2B, 3E-H)
Description: Colonies grew slowly at 25℃ and reached 47, 46, 45, 30, 49, and 45 mm on CMD, MEA, OA, PDA, V8A, and YPDA, respectively, 7 d after inoculation. The colony was colored hyaline with a smooth aerial mycelial surface on CMDA and MEA, creamy white with a fluffy aerial mycelial surface on OA, light yellow with dense aerial mycelia on PDA, translucent white with cottony aerial mycelia on V8A, and creamy white with short aerial mycelia on YPDA. Sporangia were hyaline, many-spored, and 21.3-41.2 µm in diameter (x=30.8±5.72 µm, n=15). Sporangiospores were hyaline in color, ellipsoidto-cylindrical, and 3.5-10.9 µm×2.6-8.1 μm (x=8.1±1.57 µm×5.6±0.99 μm, length/width (L/W) ratio=1.44, n=50). Chlamydospores are globose, thick-walled with irregular appendage, and 13.2-32.4 µm in diameter (x=21.9±6.74 µm, n=15).
Habitat: Filtered freshwater from a stream
Specimen examined: Yangji-ri, Goro-myeon, Gunwi-gun, Gyeongsangbuk-do, Republic of Korea, 16 Mar 2018, NNIBRFG5579, Nakdonggang National Institute of Biological Resources (collected by Namil Chung).
Note: Other strains, such as L. exigua, were mainly isolated from agricultural soil in Spain, India, and Germany [15]. In this study, NNIBRFG5579 was isolated from filtered freshwater streams.
Fig. 2
Mycelial growth on CMDA, MEA, PDA, OA, V8A, and YPDA for 7 days at 25℃. (A) Dissophora globulifera strain NNIBRFG26655. (B) Linnemannia exigua strain NNIBRFG5579. (C) Mortierella rishikesha strain NNIBRFG6579. (D) Umbelopsis autotrophica strain NNIBRFG4148. CMDA, corn meal dextrose agar; MEA, malt extract agar; PDA, potato dextrose agar; OA, oatmeal agar; V8A, V8-juice agar; YPDA, yeast extract peptone dextrose agar.
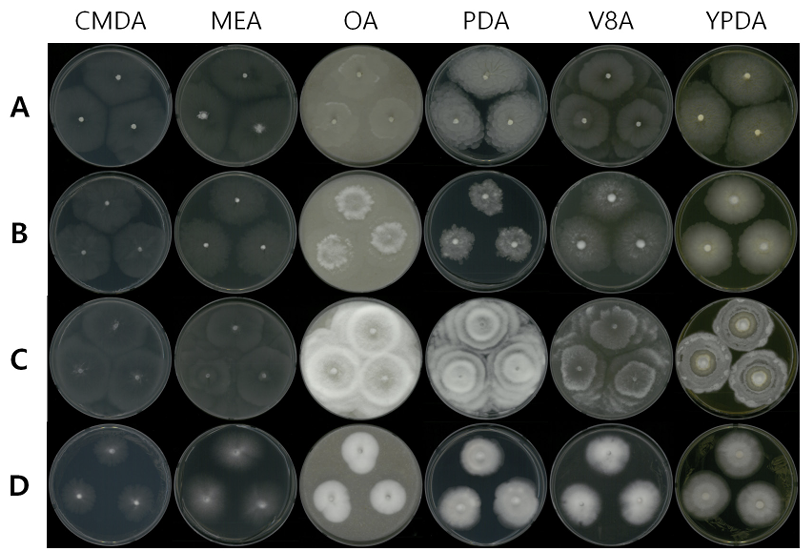
Fig. 3
Microscopic observation. A-D, sporangiophore (A-B), sporangiospore (C) and chlamydospore (D) morphology of Dissophora globulifera strain NNIBRFG26655; E-H, sporangiophore (E-G) and sporangiospore (H) morphology of Linnemannia exigua strain NNIBRFG5579; I-N, sporangiophore (I-M) and sporangiospore (N) morphology of Mortierella rishikesha strain NNIBRFG6579; O-R, sporangiophore (O-Q) and sporangiospore (R) morphology of Umbelopsis autotrophica strain NNIBRFG4148. Scale bars are 10 μm (B-D, F-H, L-N, P-R), 50 μm (K), and 100 μm (A, E, I, J, O).

Mortierella rishikesha B.S. Mehrotra & B.R. Mehrotra, ZentBl. Bakt. ParasitKde, Abt. 2: 184 (1964) [MB#334512] (Fig. 2C, 3I-N)
Description: Colonies grew fast at 25℃ and reached 52, 65, 71, 72, 69, and 48 mm on CMD, MEA, OA, PDA, V8A, and YPDA, respectively, 7 d after inoculation. The colony was hyaline with a smooth aerial mycelial surface on CMDA and MEA, creamy white with a fluffy aerial mycelial surface showing a concentric pattern on OA and PDA, translucent white with cottony aerial mycelia showing a concentric pattern on V8A, and creamy white at the margin with aerial mycelia on YPDA. Sporangia were hyaline, many-spored, and 21.8-37.7 µm in diameter (x=31.3±5.51 µm, n=20). Sporangiospores were hyaline, globose-to-oval, and 5.1-13.6 µm×3.6-8.8 μm (x=9.6±1.91 µm×6.1±1.17 μm, L/W ratio=1.57, n=50). No chlamydospores were observed in this study.
Habitat: Sediment of a stream
Specimen examined: Neunggang-ri, Susan-myeon, Jecheon-si, Chungcheongbuk-do, 27 July, 2018, NNIBRFG6579, Nakdonggang National Institute of Biological Resources (collected by Jaeduk Goh).
Note: M. rishikesha was first isolated from the forest soil in India [15]. The lack of literal information, morphological characteristics, and additional sequences (EF1) of NNIBRFG6579 were directly compared with M. rishikesha CBS 652.68.
Umbelopsis autotrophica (E.H. Evans) W. Gams, Mycological Research 107 (3): 349 (2003) [MB#373417] (Fig. 2D, 3E-F)
Description: Colonies grew slightly slow at 25℃ and reached 28, 37, 31, 37, 35, and 38 mm on CMDA, MEA, OA, PDA, V8A, and YPDA, respectively, after 7 d of inoculation. The colonies were hyaline with a smooth aerial mycelial surface on CMDA, hyaline-to-white with a cottony aerial mycelial surface on MEA, creamy white with dense aerial mycelia on OA, white-to-light pink with fluffy aerial mycelia on PDA, white with cottony aerial mycelia on V8A, and white with fluffy aerial mycelia on YPDA. Sporangia were light pink-to-vinaceous, many-spored, globose, with columellae, and 15.2-25.8 µm in diameter (x=19.7± 2.53 µm, n=50). Sporangiospores were hyaline-to-light pink, globose, and 5.1-13.6 µm×3.6-8.8 μm (x=9.6 ±1.91 µm×6.1±1.17 μm, L/W ratio=1.57, n=50).
Habitat: Sediment of a freshwater source
Specimen examined: Ocheon-ri, Yeonil-eup, Nam-gu, Pohang-si, Gyeongsangbuk-do, Republic of Korea, 25 May 2017, NNIBRFG4148, Nakdonggang National Institute of Biological Resources (collected by Hye Yeon Mun).
Note: U. autotrophica was first isolated from forest soil crumbs, root segments, and other plant material [19]. In this study, one strain was isolated from freshwater sediment.