INTRODUCTION
Grapevine (Vitis vinifera) is a widely grown fruit crop with various uses, including consumption as fresh fruit, production of jams, traditional medicine, and as raw material for juice and wine production [1,2]. Grape cultivation is socially and economically important in South Korea [3]. However, fungal diseases, particularly downy mildew, pose significant threats to commercially important grape cultivars and hybrids, resulting in substantial economic losses [4]. Oomycetes, a group of plant pathogens related to diatoms and brown algae, encompass approximately 800 species of downy mildew and over 120 species of Phytophthora [4]. Grapevine downy mildew is caused by the oomycete Plasmopara viticola and is more prevalent in regions with frequent rainfall and moderate temperatures [5].
P. viticola was initially reported in Vitis ficifolia by Choi et al. in 2017 [6] and in Vitis cognetiae by Kim et al. in 2019 in Korea [7]. However, the occurrence of this pathogen in cultivated Vitis vinifera and the methodology for growing and maintaining the pathogen under laboratory conditions require detailed reporting. Therefore, a more comprehensive identification and analysis of the behavioral characteristics of this pathogen during infection are necessary to understand its biological interactions with the host. Such comprehensive characterization holds great implications for discovering resistance genes in locally grown genotypes that can be introduced into susceptible cultivated cultivars through breeding programs [8].
Previous studies have characterized P. viticola isolates from other countries; however, the genome organization of Korean isolates has not yet been reported [9-11]. Traditional characterization methods for fungal pathogens pose challenges for P. viticola because of its biotrophic nature and inability to be cultured in artificial media. Multiple genealogy analyses using four genomic regions (internal transcribed spacer 1 [ITS1], actin [ACT], large subunit [LSU], and beta tubulin [TUB]) have been recommended for the identification of filamentous pathogens such as grapevine downy mildew. This approach has been successfully applied to characterize P. viticola populations in China and France [12,13].
In this study, 11 Korean P. viticola isolates collected from four grape-growing regions were characterized phenotypically and molecularly. The objectives of this study were as follows: (i) morphological and molecular identification of P. viticola and evaluation of its development during the course of infection; (ii) evaluation of the pathogenicity of South Korean P. viticola isolates based on their phenotypic expression in susceptible grape cultivars and the degree of host grape resistance to downy mildew infection; (iii) determination of evolutionary relationships among Korean P. viticola isolates using phylogenetic analysis. The findings of this study are crucial for understanding the pathogen's behavior during infection, which will aid in developing resistance strategies for grape cultivars. This, in turn, can help reduce the excessive use of chemical fungicides currently employed for the control of grapevine downy mildew.MATERIALS AND METHODS
Plant material preparation
Grapevine cultivars and hybrids used to isolate, maintain, and inoculate P. viticola sporangia were prepared from cuttings collected from Jeonju by the Rural Development Administration of South Korea (RDA). Rooting of the cuttings was accomplished following a previously developed procedure [14] with some modifications. Tissue-cultured hybrid plants used for sporangia maintenance were potted and maintained under the same growing conditions as those mentioned above in a greenhouse in Chungnam National University.
Collection of infected plant materials
Eleven P. viticola isolates were collected from four different grape cultivation areas: four from the RDA farm in Jeonju (JN-1, JN-2, JN-5, and JN-9), three from Cheonan (CHKS-1, CHKU-3, and CHSU-2), two from Gimcheon (KNK-3 and KNS-1), and two from Daejeon (DN-1 and DN-2) (Table 1).
The leaves of infected grapevines were collected based on the visual symptoms of grapevine downy mildew. Leaves with whitish downy sporulation on the abaxial side and oily spots on the adaxial side were collected. They were kept in a humid plastic container, checked for downy mildew infection under a light microscope, and incubated in a moist chamber with 95% relative humidity at 20℃ to stimulate active sporulation and production of fresh sporangia for spore isolation.
Isolation and maintaining of the sporangia
Isolation of P. viticola sporangia was conducted according to the procedure developed by [15]. Briefly, fresh sporangia were collected from a single oily spot on infected leaves, suspended in 1ml sterilized distilled water in plastic vials mixed thoroughly and diluted to obtain sporangia suspension of 4×104 sporangia/mL. Healthy grape leaves used for leaf disc preparation were surface sterilized in 1% sodium hypochrolite for 2 minutes followed by three time wash in sterile water; 2 cm in diameter leaf discs for both propagation and spore isolation were excised from the 3rd to 6th leaves counted from the shoot tips using a sterilized cork borer.
Well prepared Petri plates with moist filter paper to keep relative humidity to around 100% in the medium were used as medium for spore isolation; the prepared 10 leaf discs for Vitis vinifera cv Italia and Hongju×Hucbarad (hybrid) each, were gently perforated using a sterile piercing equipment, then placed on the wet filter paper abaxial side up and inoculated with drops of 50 µL sporangia suspension with 4 ×104 sporangia/mL concentration, Samples with inoculated leaf discs were kept at 20℃ for 24 hours in darkness; after infection samples were incubated at 20℃ under 16 hours photoperiod. Sporangia produced on each leaf discs were again isolated thrice to insure purity of the strains and then propagated weekly on leaf discs following the above detailed procedure.
Pathogenicity test
Eleven P. viticola isolates were inoculated onto grapevine leaf discs to evaluate their pathogenicity. A 50-mL suspension of 5×104 sporangia of each P. viticola isolate was used for inoculation. Samples were checked daily for sporulation, starting from the day after inoculation. Pathogenicity among the field isolates was evaluated based on three parameters: (i) latent period, defined as the number of days from inoculation until visible sporulation on the disc surface; (ii) number of leaf discs with sporulation and discs with a larger sporulation area than the inoculated site, measured 8 days after inoculation; and (iii) diameter of sporulation (in mm) measured 10 days after inoculation; the experiment was repeated three times to ensure reliability of pathogenicity among isolates.
Grapevine host resistance to downy mildew infection
Screening for grape host resistance to downy mildew infection was conducted with 3 independent experimental assays performed on 845 cultivars. These cultivars were collected and inoculated with the most virulent isolate, JN-9, which was selected during pathogenicity testing. The degree of host resistance among the tested grape cultivars was recorded 8-10 days after inoculation. The disease scaling method developed by OIV-452 (Organisation Internationale de la vigne et du vin) was adopted to evaluate cultivar resistance based on the presence of downy mildew patches on the leaf surface following infection.
Phylogenetic analysis
Leaf discs infected with P. viticola isolates were collected after sporulation on the surface and lyophilized in liquid nitrogen for DNA extraction using the CTAB method, as described by Tripathy et al. [16]. Four different DNA regions, namely the internal transcribed spacer 1 (ITS1), a fragment of cytochrome c oxidase II (COX2) gene, beta tubulin (TUB), and actin (ACT), were selected for molecular characterization of the isolates (Table 2). Phylogenetic relationships among the field isolates were analyzed using the maximum likelihood (ML) method based on the Kamura 2-parameter model with 100 non-parametric bootstraps in the Mega 7 software. The number of conserved sequence sites, variable sites, and parsimony-informative sites were determined.
Statistical analysis
SAS (Statistical Analysis System 9.1, SAS Institute, Cary, NC, USA) was used for all statistical analyses. One-way analysis of variance (ANOVA) was used to statistically analyze the results. p<0.05 was considered statistically significant. Tukey’s test was used for comparisons (p<0.05) when variance was observed among the groups.
Leaf surface sporulation method developed for in vitro Plasmopara viticola inoculation
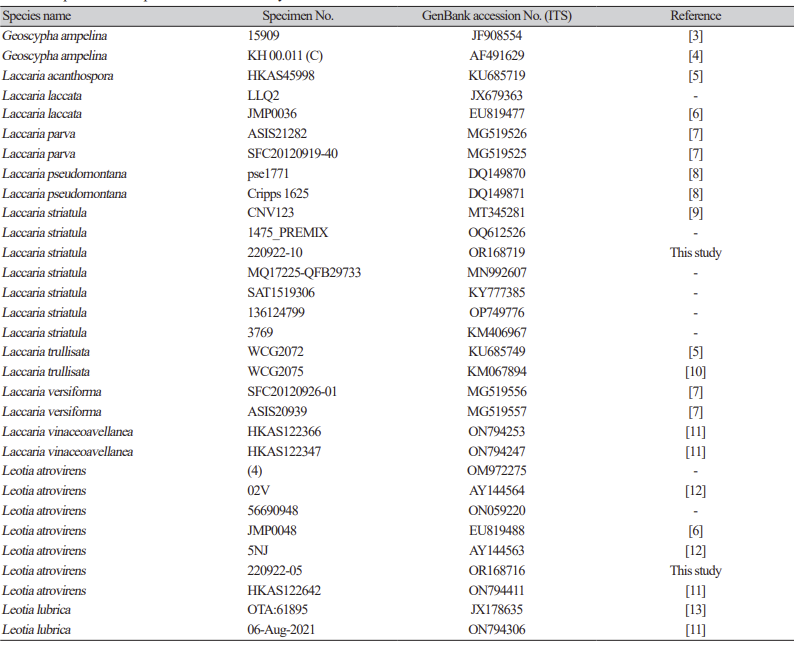
In this study, in vitro inoculation of P. viticola sporangia was performed on leaf discs obtained from susceptible grapevine cultivars, Vitis vinifera ʻItalia’ and the hybrid Hongju×Hucbarad. Sporangia released zoospores after approximately three hours at room temperature. Uniform round zoospores were observed swimming in suspension under a light microscope (Fig. 1B). Germ tubes formed by the zoospores were visible on the surface of the leaf discs, and their size and number increased within 24-48 hours after inoculation for all the cultivars. (Fig. 1C and D).
Four days after infection, fresh whitish sporangiophores and sporangia on the surface of the discs were visible to the naked eye. Additionally, yellowish patches were observed on the adaxial side of the leaves, as reported by Kennelly et al. [17]. All inoculated isolates showed varying degrees of sporulation ranging from abundant and profuse to weak mycerial growth on the surface of two Vitis cultivars, (Fig. 1E and F).
Fig. 1
Phenotypic characterization of Plasmopara viticola by leaf disc method. (A) Lemon shaped sporangia. (B) Zoospores released from mature sporangia 3-6 hours of incubation at room temperature in suspension. (C and D) Development of a germ tube ready to penetrate through stomata; Surface sporulation on the disc at four days after infection on two different cultivars. (E) Vitis vinifera cv Italia (F) Vitis vinifera cv Hongju×Hucbarad (hybrid). Scale bar=2 mm.
Phylogenetic relationships among Korean P. viticola isolates
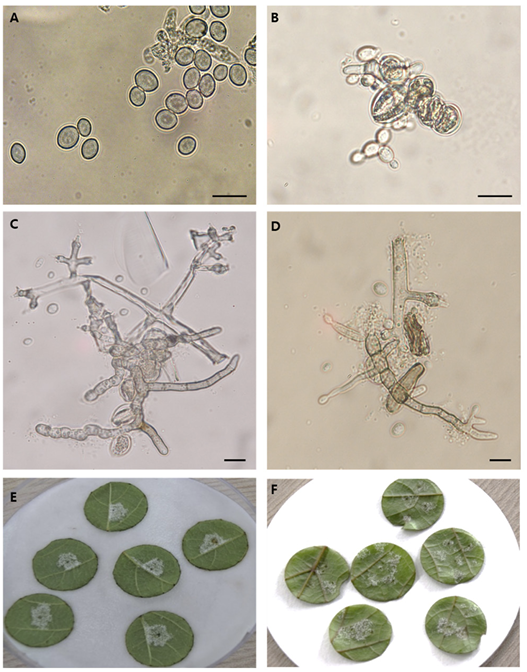
The sequences of four genomic regions from 11 Korean isolates and selected European, American, and Chinese isolates were used to determine the phylogenetic relationships among the studied South Korean P. viticola isolates. Phylogenetic trees for all fragments of the four genomic regions are shown in Fig. 2. The alignment of all DNA regions studied consisted of 14 nucleotide sequences, with lengths of 278 bp for ITS1, 540 bp for actin, 543 bp for β-tubulin, and 631 bp for cytochrome c oxidase II. The aligned sequences were trimmed to remove gaps, resulting in sequences with evolutionary relationship parameters as shown in Table 3. All aforementioned sequences for the four genomic regions were concatenated to create a consensus tree for phylogenetic species recognition, following the method by Taylor et al. [18]. The 14 concatenated nucleotide sequences were aligned in MEGA7, resulting in 1,705 nucleotide sites representing the four DNA fragments. Phylogenetic analyses of these genes separately confirmed the evolutionary relationships observed in each region (Fig. 2). The 11 isolates collected from four different geographical regions of South Korea clustered together in a common monophyletic lineage, indicating a shared, most recent common ancestor. When comparing the degree of identity with foreign P. viticola isolates, our isolates showed a closer relationship with the Chinese P. viticola isolate (accession number KF220645) available in the NCBI database than with European (JF897799) and American (MK358364 and MT017599) P. viticola isolates. However, the Korean isolates did not show any geographically based genetic diversity.
Fig. 2
Molecular phylogenetic tree obtained by linking Plasmopara viticola Actin, Cytochrome C oxidase II, Internal Transcribed Space1 and β-Tubulin by maximum likelihood. Sequences were downloaded from National Center of Biotechnology Information database. The tree was constructed with 100 bootstrap values and the proportion by which associated haplotypes are grouped together are presented next to the branches. Evolutionary analysis was performed using MEGA7.
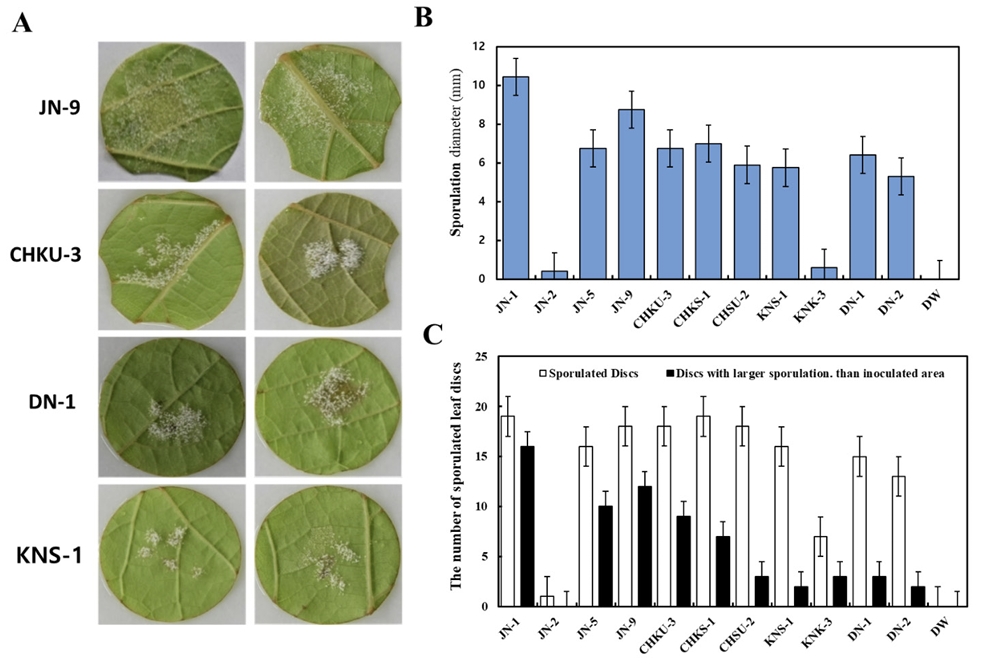
Pathogenic variability of 11 P. viticola isolates
Spores from 11 P. viticola isolates collected from four viticultural regions were inoculated into Italian grape cultivars to evaluate their pathogenicity. The JN-9 strain from the Jeonju area exhibited visible sporulation within four days of inoculation and had the shortest incubation period compared to the other strains (5-6 days). The isolates showed varying sporulation phenotypes, with JN-2 displaying weak sporulation, and JN-1 and JN-9 demonstrating high sporulation capacity (Fig. 3A).
The degree of sporulation was evaluated by analyzing the size of the sporangium patches on the eighth day postinfection. JN-1 exhibited the highest degree of sporulation, whereas other isolates, including KNS-1, showed smaller sporulation areas. Spore diameters also varied among the isolates, with JN-1 having the largest average diameter and JN-2 having the smallest (Fig. 3B and C).
The degree of sporulation was evaluated by analyzing the size of the sporangium patches on the eighth day postinfection. JN-1 exhibited the highest degree of sporulation, whereas other isolates, including KNS-1, showed smaller sporulation areas. Spore diameters also varied among the isolates, with JN-1 having the largest average diameter and JN-2 having the smallest (Fig. 3B and C).
Fig. 3
Pathogenicity variability among field isolates (Plasmopara viticola) based on their sporulation capacity on the leaf discs. (A) Selected most virulent isolates from four grape cultivation regions in Korea. (B) Sporulation potential exhibited by field isolates. (C) Average sporulated leaf discs for the tested isolates.
Host resistance screening in Korean cultivars
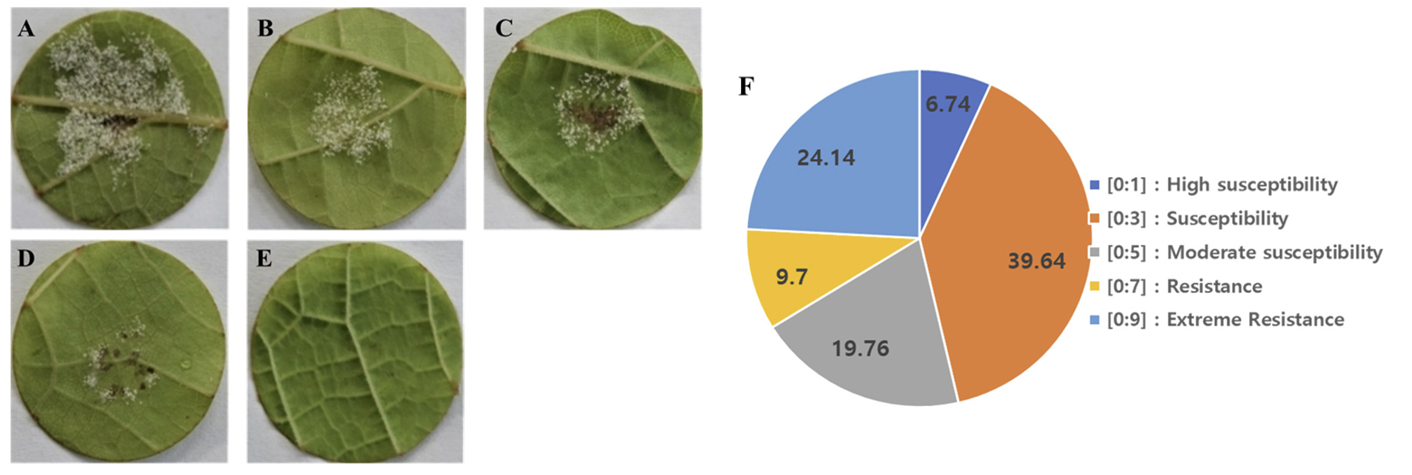
To evaluate the degree of resistance of grapevine cultivars to downy mildew infection, 845 cultivars were collected and inoculated with isolate JN-9. All the collected healthy leaves were surface sterilized in 1% sodium hypochlorite solution and prepared in 90×15 mm petri plates in the same procedure as detailed above for the spore isolation assay. 10 leaf discs were used for each petri plate replicated three times; well prepared leaf discs of the 4th leaves for Each cultivar counted from the apical bud were inoculated with 6×104 sporangia/ml suspension of P. viticola isolates. Inoculated leaf discs were kept at 20℃ for 24hrs in absence of light; then incubated at 16hours photoperiod. The degree of host resistance among grape cultivars tested were recorded at 10 days’ post inoculation by adopting disease scaling developed by OIV452 grouping them into five categories, ranging from category A, representing highly susceptible cultivars (scale [0:1]), to category E, representing extremely resistant cultivars (scale [0:9]) (Fig. 4A and E).
Of the 845 tested cultivars, 7% showed abundant and unrestricted sporulation covering almost the entire leaf disc; these were classified in category A (scale [0:1]) as highly susceptible cultivars. Category B, which comprised the highest proportion (41%), represented susceptible cultivars that allowed abundant sporulation but were limited to inoculation areas without necrosis (scale [0:3]). Approximately 20% of the cultivars exhibited moderate susceptibility, which was characterized by limited sporulation and weak necrotic spots (scale [0:5]). Additionally, 8% of the cultivars had restricted sporulation to some extent and exhibited necrotic spots, placing them in category D as resistant grapevine cultivars (scale [0:7]). Notably, 24% of the cultivars used for screening showed no symptoms of downy mildew disease-neither sporulation nor necrotic spots were observed on the surface of any of the 10 tested leaf discs; these cultivars were classified as category E, extremely resistant cultivars (scale [0:9]) (Fig. 4F).
Fig. 4
Grape host resistance reaction based on sporulation phenotype on the leaf discs against Plasmopara viticola JN-9. (A) abundant sporulation no necrotic spot (very susceptible). (B) Limited sporulation no necrotic spot (susceptible). (C) Scattered sporulation with weak necrotic flecks (moderately susceptible), (D) Rare sporulation and strong necrotic spot (resistant) neither sporulation nor necrosis extremely resistant, (F) The proportion of grape cultivars with their respective Degree of resistance to downy mildew infection. The images were taken at 10 days after inoculation.
DISCUSSION
Plasmopara viticola is an oomycete that causes severe damage to vineyards worldwide [19]. Grapevine downy mildew is primarily established through infection by activated oospores that germinate on the leaf surface, penetrate the stomata, and develop hyphae, forming mycelial structures within the mesophyll cells. Colonization involves the development of globose hostoria that absorb nutrients from the host and secrete biomolecules for colonization [20,21]. Visible symptoms of downy mildew include oily spots on the upper leaf surface, whitish mycelial structures protruding through the stomata, and necrotic lesions that can lead to defoliation [17,22]. Primary infection with resting oospores is followed by a cycle of asexual zoospore production. Sporangia isolated from leaves produce active zoospores within three hours. Under favorable conditions, zoospores encyst upon reaching the stomata and develop germ tubes for penetration. In vitro spore isolation was conducted using susceptible grape cultivars such as Vitis vinifera ʻItalia’ and the hybrid Hucbarad, which allowed abundant sporulation. Leaf discs excised from healthy vines facilitated the isolation and maintenance processes. However, laboratory-controlled conditions may not fully reflect field conditions, and further analyses should involve in vivo experiments.
Pathogenicity of P. viticola relies on the secretion of an array of biomolecules that modulate biological as well as physiological functions of the host to promote infection. These virulence characteristics are expressed and evaluated through the ability of the pathogen to produce active sporulation on the surface of the grape leaf [13]. Isolates of P. viticola showed varying degree of sporulation potential on the surface of leaf discs used in this study. Isolates JN-9 and JN-1 produced the highest sporulation compared to other tested isolates. This is in line with the findings of Gómez-Zeledón et al. [13] who showed that different strains produced diverse sporulation on grape leaf discs. The pathogenicity of Plasmopara viticola depends on isolate’s genetic makeup and varies according to environmental conditions as well as the degree of host resistance; this confirms what was reported in a study in China where isolates collected from resistant varieties from Chinese vineyard showed greater pathogenicity than isolates collected from susceptible ones regardless of geographical origin [23]. Isolates collected from the Jeonju farm exhibited high phenotypic variability in terms of sporulation capacity. This variability may be attributed to the higher mutation rate and occurrence of sexual reproduction in P. viticola, leading to diverse strains, even when collected from the same plant [23-26]. Gene-for-gene coevolution between the pathogen and host plant contributes to the development of genes that allow the pathogen to evade host defense responses [27]. Most cultivated Vitis cultivars showed varying levels of susceptibility to P. viticola infection, with a large proportion categorized as moderately to severely susceptible. However, resistant cultivars were able to restrict the sporulation of P. viticola and expressed hypersensitive cell death in the form of necrotic spots at the inoculation sites. Resistant cultivars activate defense mechanisms by recognizing pathogens and limiting infection. Some known resistance genes include Rpv11 in cv Regent and Rpv10 in cv Solaris [28]. On the other hand, susceptible cultivars lack the pathogen recognition receptor molecules making them unable to recognize the presence of pathogenic microorganisms thereby rendering them a suitable environment for the pathogen to establish the disease [29].In addition to resistance genes grape cultivars exhibiting extreme resistance often possess visible trichome structures on the leaf surface, providing physical resistance by preventing pathogen contact with stomata. The absence of downy mildew symptoms in resistant cultivars can be attributed to the physical barrier created by leaf hair. Physiological adaptations and anatomical structures of stomata in resistant cultivars also contribute to the early arrest of the infection process, even in the absence of leaf hair structures [30,31].
Phylogenetic analysis of the conserved genomic regions revealed that all 11 isolates from different geographical locations clustered together, indicating a lack of significant genetic diversity regardless of the geographical locations from which isolates were collected; this is consistent with the results published by Zhang et al. [12] Sexual reproduction and higher rate of genetic mutation were reported to be the main causes of higher level of polymorphism among P. viticola population; the presence of two mating types for this pathogen explains diversity resulting from sexually produced oospores [32]. In our case a little genetic diversity was observed among isolates implying the absence of significant genetic diversity. Moreover, all isolates were collected in a period of higher rainfall accompanied with humid and moderate temperature; favorable condition for the asexual reproduction of grape downy mildew causing agent resulting in identical progenies. This study demonstrates the successful growth and varying pathogenicity of P. viticola isolates on leaf discs from the susceptible cultivars. Tested Vitis cultivars exhibited different levels of susceptibility to pathogenic infections. Phylogenetic analysis revealed a common evolutionary relationship among Korean isolates, irrespective of their geographical origin. These findings provide valuable information for future studies on plant-pathogen interactions and highlight the limited impact of geographical location on the evolutionary potential of the pathogen.