INTRODUCTION
Tan spot, caused by Pyrenophora tritici-repentis (Died.) Drechs., is a major disease affecting wheat worldwide [1]. Since its first report in the United States, the disease incidence and severity have been increasing in wheat grown worldwide [2,3]. Although the plant pathogen has primarily been isolated from wheat, it has also been reported to have a relatively broad host range, including rye, barley, and numerous grass species [4,5].
The pathogen P. tritici-repentis can survive saprophytically on infected wheat stubble and plant residues [6]. The infection begins when ascospores infect young wheat seedling leaves and produce lesions. These lesions are small tan-brown spots with a yellow halo that later expand [7]. As the disease progresses, tan spot lesions gradually merge, resulting in a substantial area of necrotic tissue [8]. Sporulation in Pyrenophora species plays an important role in the morphological characterization of conidia, aiding in the identification of isolates and subsequent research [9].
Previous studies have reported that this pathogen causes 20-50% of severe yield losses in wheat production [10,11]. Various fungicides have been reported to be effective in controlling P. tritici-repentis. These include azoxystrobin and trifloxystrolin, which inhibit respiration and energy production in fungal cells; iprodione, which inhibits signal transduction; mancozeb, which inhibits multi-site contact activity; and cyproconazole, tebuconazole, flutriafol, flusilazole, epoxyconazole, difenoconazole, and propiconazole, all of which inhibit sterol biosynthesis in the membrane [12]. Azoxystrobin and trifloxystrobin belong to the quinone outside inhibitor (QoI) fungicides [13].
In April 2021, numerous tan spot lesions were observed in commercial wheat fields, and potential causal pathogens were isolated from these lesions. The objectives of this study were to identify the fungal pathogens responsible for tan spot disease in wheat and select an appropriate fungicide to control these pathogens. Until now, scant information has been available on P. tritici-repentis isolates present in wheat in Korea.
MATERIALS AND METHODS
Collection and isolation of the fungal isolates
The samples were collected from wheat leaves in Suncheon, Korea (34°55'22.13"N, 127°29'46.31"E). The infected leaf tissue was sectioned (5×5 mm), surface-sterilized by dipping in 1% sodium hypochlorite for 30 s, followed by 70% ethanol for an additional 30 s. Subsequently, the samples were rinsed thrice with sterile water and placed on water agar medium supplemented with streptomycin (100 μg/mL). From each sectioned leaf, a single colony was selected and transferred to hyphae emerging from the samples on V8potato dextrose agar (PDA; Difco, Detroit, MI, USA) medium (10 g of PDA 150 mL of V8 Juice; 3 g of CaCO3 ; and 15 g of agar per liter). Subsequently, the obtained cultures were preserved in a 20% glycerol stock at -80℃ for further study [14]. Notably, a representative isolate has been deposited in the Korean Agricultural Culture Collection (accession no. KACC 410061).
Morphological characterization
The cultural characteristics of the isolates were examined by cultivating them on PDA medium in the dark at 25℃ for 7 days. To induce conidia, wheat stems and seeds were used as substrates on water agar medium (1.5% agar). Water agar, inoculated with sterilized wheat stems or seeds, was incubated under a 12hour light and dark cycle for 10 days. For microscopic characterization, the isolates were examined using a Zeiss Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany) with an Axiocam 305 color microscope camera and Zeiss ZEN 2.6 Software (Carl Zeiss, Jena, Germany).
Molecular analysis
Genomic DNA was isolated using the NucleoSpin® Plant II kit (Macherey-Nagel, Duren, Germany), following the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed with 20 ng of DNA and 10 pmol of each primer (Table 1), utilizing the i-StarMAX II PCR master mix system (iNtRON Biotechnology Inc., Seongnam, Korea). The amplification conditions were as follows: 94℃ for 3 min, followed by 30 cycles of 95℃ for 30 s, 55℃ for 1 min, and 72℃ for 30 s. Sequence information was obtained from Bioneer Corporation (Bioneer, Daejeon, Korea). The obtained sequences were deposited in the GenBank database, managed by the National Center for Biotechnology Information (NCBI).
Reference genes for the phylogenetic analysis were obtained from the NCBI database and are listed in Table 2. The sequences of genes, including internal transcribed spacer (ITS), largest subunit ribosomal RNA (LSU), glyceraldehyde-3-phosphate dehydrogenase (gapdh), and RNA polymerase II secondlargest subunit (rpb2), were individually aligned for each gene region using the multiple sequence alignment program MUSCLE (multiple sequence comparison by log-expectation) [15]. Subsequently, the aligned sequences were combined to generate multi-locus datasets using the MEGA 11 software [16]. A phylogenetic tree was then constructed using a concatenated dataset and the maximum likelihood method [17], employing the Kimura 2-parameter model and 1,000 bootstrap values [18]. Branch lengths indicate the amount of change considered to have occurred between two nodes.
Pathogenicity test
Pathogenicity tests were conducted on three biological replicates of the wheat cultivar “Geumgang.” The plants were grown in 0.7-liter pots at 20℃ under a 12-hour light cycle for 3 weeks in duplicate. To prepare the inoculum, fungal mycelium was cultured in PDA medium for one week. The freshly cultivated mycelium was macerated with 10 mL of sterilized distilled water containing 0.1% Tween 20 using an ULTRA-TURRAX® homogenizer (IKA-T10, Guangzhou, China), resulting in homogenized mycelial suspensions. The prepared suspension was evenly sprayed over the entire surface of the plant leaves. The control group was sprayed with 0.1% Tween 20. The infected plants were then incubated at 20℃ in the dark with 100% relative humidity in plastic bags for 3 days. Subsequently, they were transferred to a growth chamber under a 12-hour light cycle with high humidity until the disease developed.
in vitro selection of fungicides
To select an appropriate fungicide against P. tritici-repentis on wheat, mycelial agar plugs (6-mm in diameter) were collected from the margins of the mycelia on PDA. These plugs were then placed upside down on V8 Juice agar medium (80 mL of V8 Juice, 15 g of agar, pH 7.0) supplemented with fungicides at different concentrations. After incubation at 25℃, the inhibition of mycelial growth was measured after 2 weeks. The fungicides were applied at the same amount as the commercially recommended concentration for controlling tan spot disease in barley. The following fungicide concentrations were used: azoxystrobin (110 μg/mL), iprodione (250 μg/mL), mancozeb (1,125 μg/mL), and tebuconazole (125 μg/mL). The inhibition of mycelial growth was calculated using the following formula: (1–[colony diameter on V8 Juice agar medium with each fungicide/colony diameter on V8 medium without the fungicide])×100. Statistical analyses were performed using the SPSS software version 18.0 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
Collection of fungal isolates
In April 2021, the incidence of tan spot disease on wheat leaves in commercial fields in Suncheon was approximately 5%. The typical symptoms included oval to irregularly shaped tan-brown spots with a yellow halo on the leaves (Fig. 1A and B). Three representative isolates confirmed to cause tan spot, namely, KACC 410061 (SYP-F0555), SYP-F0557, and SYP-F0558, were characterized to determine their identity. In contrast to occurrences in other previously reported countries [10,11], our results showed that wheat grown in Korea was not significantly affected by tan spot disease. However, potential future impacts, such as climate change, should also be considered.
Fig. 1
Disease symptoms on the leaves of naturally infected Triticum aestivum, morphological characteristics of Pyrenophora tritici-repentis, and pathogenicity. (A) A typical symptom of tan spot caused by P. tritici-repentis on wheat leaves. (B) The photographs, from left to right, show the progressive severity of tan spot disease on wheat leaves. The (C) front and (D) reverse sides of a 2-week-old colony on potato dextrose agar. (E) Pseudothecia formed on the wheat stem after 4 weeks. (F) Immature pseudothecium under microscopy. (G) Conidia of P. tritici-repentis. (H) Results of pathogenicity tests. Plants were inoculated with sterile distilled water (left) and a macerated mycelial suspension of P. tritici-repentis at 7 days post-inoculation (dpi). (I) Typical disease symptoms of tan spot disease after 7 dpi. (H) Close-up view of white circle in (I).

Morphological characteristics
The isolates exhibited irregular dense mycelial growth on PDA, and the mycelia were dark greenish-gray with fluffy aerial hyphae. The colonies showed a dark olive color when observed from the underside of the plate (Fig. 1C and D). These morphological characteristics are consistent with previously described data for P. tritici-repentis [23].
To induce conidial production, we adapted the optimized sporulation method described by Lamari and Bernier [10] and Jacques et al. [14]. Following the former method, the strain was cultured in V8-PDA, and sterilized distilled water was added to the culture. Hyphae were flattened with a sterilized spreader and incubated at 15℃ to obtain conidia. In contrast, the latter method involves transferring the strain cultured on V8-PDA to near-ultraviolet (UV) light and inducing conidia in a dark room at 15℃.
Despite applying several protocols, we failed to induce sexual or asexual conidia in this study. Several studies have also reported difficulties in inducing conidia [9,24]. In this investigation, sterilized wheat stems and seeds were placed on water agar medium. Consequently, we induced the formation of pseudothecia (Fig. 1E and F) [9,25,26] and conidia (Fig. 1G). The generated conidia exhibited cylindrical, straight or slightly bent shapes, a rounded apex with 5 to 10 septa, and dimensions of 46.6-213.4×3.9-12.8 μM (n=20). The observed morphological characteristics were consistent with those of P. tritici-repentis (Fig. 1). G) (8,14). A previous study [27] reported that pseudothecia mature when placed in a dark condition at 5℃ for 30 days. Although we did not subject them to these specific conditions, pseudothecia were observed.
Molecular identification of the isolates
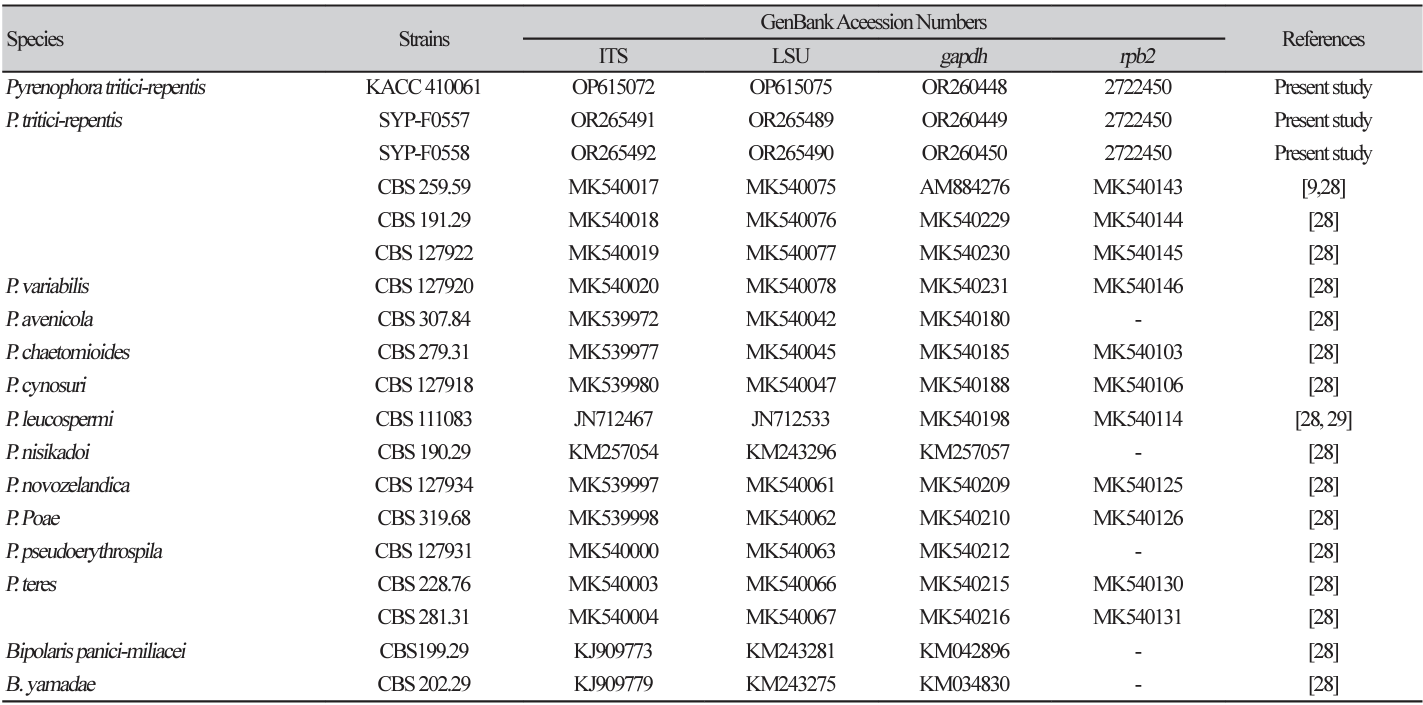
Because complete observation of sexual and asexual conidia was not achieved, a more detailed phylogenetic analysis was conducted. The ITS, LSU, gapdh, and rpb2 gene sequences matched those of P. tritici-repentis strain CBS 259.59 (accession nos. MK540017 (100%), MK540075 (99.88%), and AM884276 (100%), respectively). The rpb2 sequences showed a similarity of 93.85% to those of P. triticirepentis strain MAFF:511122 (LC685473). The resulting sequences have been deposited in GenBank (Table 2). The results of sequencing analysis clearly indicated that all representative isolates were P. tritici-repentis.
In the phylogenetic tree, the ITS, LSU, gapdh, and rpb2 alignments included three representative isolates, 13 authenticated reference sequences, and two Bipolaris sp. as outgroups (Table 2). Phylogenetic analysis was conducted using the MEGA X program based on concatenated datasets of these four multigenes. The maximum likelihood estimation revealed that the three representative isolates, KACC 410061, SYP-F0557, and SYP-F0558, clustered in the same clade as P. tritici-repentis CBS 191.29, CBS 127922, and CBS 259.59, respectively, all with 100% bootstrap values (Fig. 2A). The phylogenetic analysis robustly supported the identity of the representative isolates as P. tritici-repentis.
Molecular identification of the isolates
Because complete observation of sexual and asexual conidia was not achieved, a more detailed phylogenetic analysis was conducted. The ITS, LSU, gapdh, and rpb2 gene sequences matched those of P. tritici-repentis strain CBS 259.59 (accession nos. MK540017 (100%), MK540075 (99.88%), and AM884276 (100%), respectively). The rpb2 sequences showed a similarity of 93.85% to those of P. triticirepentis strain MAFF:511122 (LC685473). The resulting sequences have been deposited in GenBank (Table 2). The results of sequencing analysis clearly indicated that all representative isolates were P. tritici-repentis.
Fig. 2
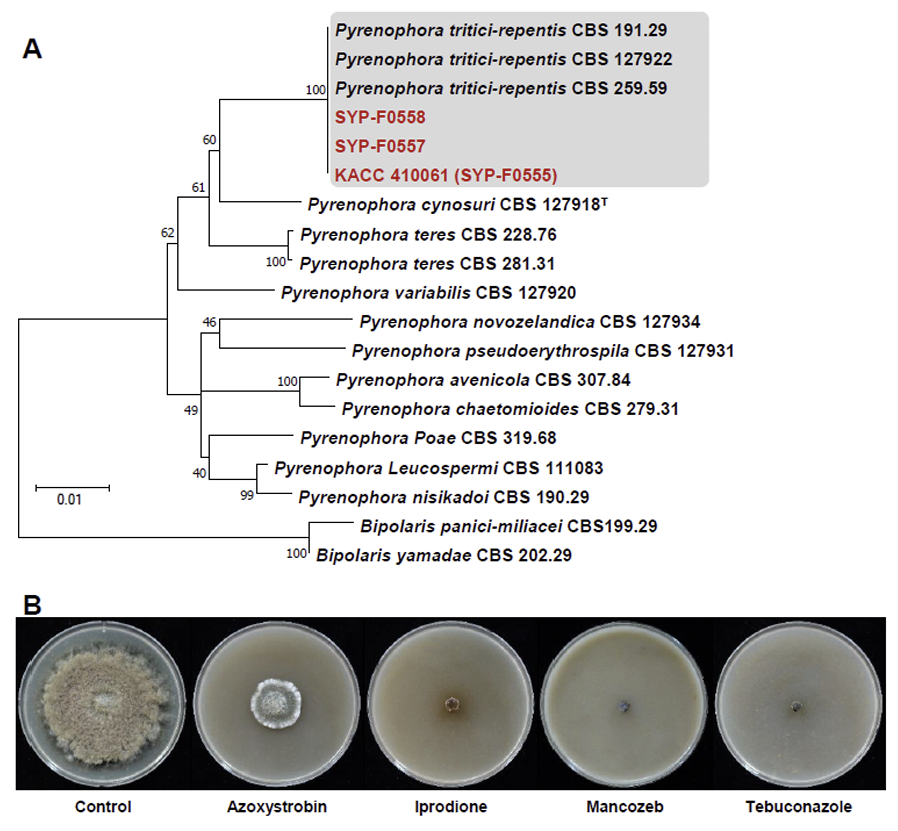
Phylogenetic analysis and inhibition zone of Pyrenophora tritici-repentis by four fungicides. (A) A maximum likelihood (ML) tree was generated from concatenated ITS, LSU, gapdh and rpb2 sequences from 16 strains representing Pyrenophora species, including isolates KACC 410061 (SYP-555), SYP-F0558, and F0557. Clades containing P. tritici-repentis and present isolates are shaded, in which existing P. triticirepentis isolates and present isolate are shown in bold in black and red, respectively. A bootstrap value is indicated by the number at each node. Bootstrap=1,000. (B) Inhibitory effects of four fungicides on the mycelial growth of P. tritici-repentis on V8-Juice agar medium. ITS, internal transcribed spacer; LSU, largest subunit ribosomal RNA; gapdh, glyceraldehyde-3-phosphate dehydrogenase; rpb2, RNA polymerase II secondlargest subunit.
Pathogenicity test
Initial symptoms manifested at 5 days post-inoculation (dpi), with typical tan spot symptoms becoming evident at 7 dpi. The affected wheat leaves were chlorotic, with small dark brown to black spots (Fig. 1H-J). In contrast, no symptoms were observed in plants in the control group. Pathogenicity tests were performed with three biological replicates in duplicate. The pathogen was successfully re-isolated from symptomatic tissues but not from the control group. The identity of the re-isolated pathogens was confirmed through the analysis of four genes, thus fulfilling Koch's postulates.
Fungicidal activity
In Australia, the fungicidal control of wheat involved applying azoxystrobin, iprodione, mancozeb, and tebuconazole, which resulted in a reduction in grain yield loss compared with untreated controls in the field (12). Similarly, we employed these four fungicides with different modes of action in this study.
The mycelial growth inhibition rate was calculated by comparing the growth rate of mycelia grown in the medium treated with the fungicides with that of the control. The inhibitory effects of azoxystrobin, iprodione, mancozeb, and tebuconazole on P. tritici-repentis were 72.6, 90.4, 91.8, and 91.8%, respectively. Among the tested fungicides, mancozeb (91.8%) and tebuconazole (91.8%) exhibited the highest mycelial growth inhibition rates, followed by iprodione. However, azoxystrobin, a QoI fungicide, showed the lowest mycelial growth inhibition rate (Fig. 2B). Because this was an in vitro experiment, further in vivo validation is required.
To the best of our knowledge, this is the first report of tan spot disease on wheat leaves caused by P. triticirepentis in Korea. This study represents an initial step toward enhancing wheat productivity and developing control strategies for tan spot disease on wheat leaves in Korea.