Orixa japonica Thunb., commonly known as East Asian orixa or Japanese orixa, is a dioecious shrub belonging to the family Rutaceae. It is native to China, Japan, and South Korea, and is typically found in either open or dense forests on sunny slopes [1]. Because of its cold and salty wind tolerance, it thrives in the coastal areas of Korea [2]. In Asian countries, O. japonica has historically served multiple purposes, including its utilization as an insecticide for livestock and its application in traditional medicine as a febrifuge and analgesic [3,4]. This species is renowned for its diverse quinoline alkaloids, which have exhibited several pharmacological activities [3,5].
During our routine field forays investigating the diversity of phytopathogenic fungi, the asexual morph of a previously unknown powdery mildew fungus on O. japonica plants has been found in Jeju and Wanju, Korea since 2013. Initially, the infected plants displayed circular to irregular white colonies with abundant conidiophores and conidia on the leaves and stems (Fig. 1A). As the disease progressed, powdery patches covered both sides of leaves and young stems with a thin layer of mycelia, causing leaf senescence and early defoliation (Fig. 1A). Occasionally, the upper surface of the affected leaves exhibited only leaf yellowing, as powdery mildew colonies were abundant on the lower surface (Fig. 1B). No chasmothecia were found until the infected leaves fell in December 2022. The collected samples were preserved at the Korea University Herbarium (Seoul, Korea) as KUS-F27419 (31 Jul 2013, Gyorae Recreational Forest, Jeju), F30246 (24 Oct 2017, Seongpanak Trail, Jeju), F33017 (28 Jun 2022, Sanggwan Forest, Wanju), and F33401 (18 Oct 2022, Sanggwan Forest, Wanju).
Detailed morphological characteristics of the fungus were observed using fresh samples under an Olympus BX50 microscope (Olympus, Tokyo, Japan). Photomicrographs were taken using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). The following anamorphic characteristics were examined. Hyphae were superficial, straight to wavy, branched, and 4-7 μm wide. Hyphal appressoria were multi-lobed or moderately lobed, single or in pairs, and 3-8 μm wide (Fig. 1C). Conidiophores were solitary, upright, 40-72×8-10 μm, and composed of 3(-4) cells (Fig. 1DG). Foot-cells of conidiophores were cylindrical, straight to mildly flexuous, and (12-)18-24 μm long (Fig. 1D-G). Conidia were formed singly on conidiophores, elliptical to ellipsoid˗oval, 28-42×16-22 μm with a length/width ratio of 1.5-2.1, vacuolate, and devoid of conspicuous fibrosin bodies (Fig. 1H). Germ tubes varied in shape and were produced at the perihilar position of the conidia (Fig. 1I). These characteristics were consistent with the description of Erysiphe orixae (U. Braun & Tanda) U. Braun & S. Takam. [6].
Fig. 1
Erysiphe orixae found on Orixa japonica. A: Typical powdery mildew symptoms on the leaves. B: Leaf yellowing caused by hypophyllous infection of the powdery mildew. C: Appressorium formed on a hypha. D-G: Conidiophores. Arrow in G shows the position of an abnormal septum. H: Conidia. I: Conidia in germination.

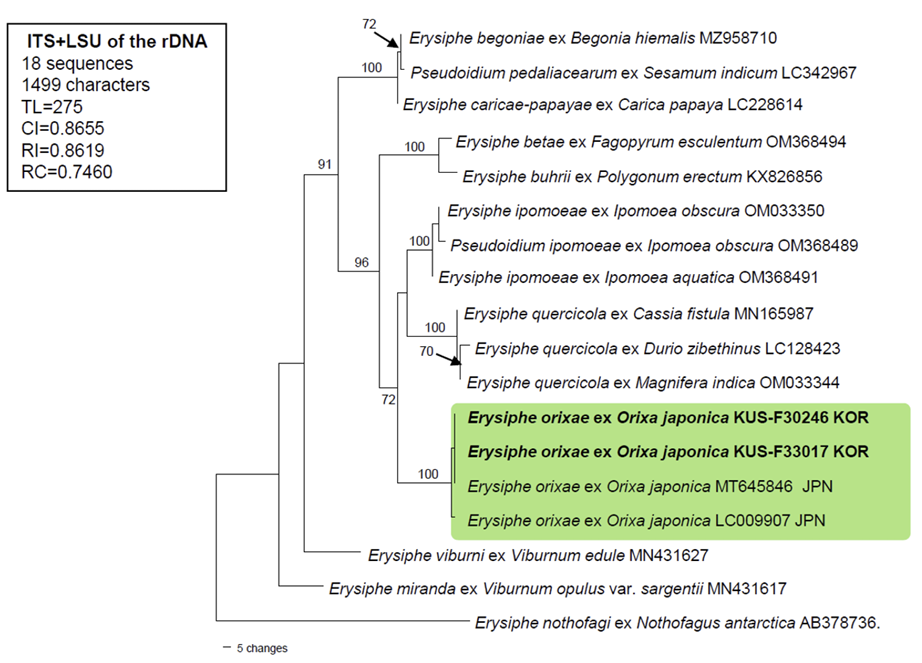
To confirm the morphology-based identification, the two aforementioned specimens (F30246 and F33017) were subjected to the molecular phylogenetic analysis. The nucleotide sequences of the internal transcribed spacer (ITS) regions, including the 5.8S gene, and the large subunit (LSU) gene of the rDNA, were determined [7], and the resulting sequences were submitted to GenBank under the accession numbers: OR527920 for ITS, and OR527922 and OR527923 for LSU. To perform the phylogenetic analysis, a combined alignment of ITS+LSU was created in MEGA11 using 18 sequences [8]. Of these, 2 new concatenated sequences of E. orixae were aligned using the MUSCLE command with 15 closely related Erysiphe sequences and 1 sequence of E. nothofagi retrieved from GenBank [8]. A phylogenetic tree was constructed in PAUP* 4.0a based on the maximum parsimony (MP) method using a heuristic search option with the “Tree-bisection-reconstruction” (TBR) algorithm, employing 100 random sequence additions to f ind the global optimum tree. All sites were treated as unordered and unweighted, and gaps were treated as missing data [9]. The robustness of the internal branches was evaluated using bootstrap (BS) analysis with 1,000 replicates. Tree scores, such as tree length, consistency index (CI), retention index (RI), and rescaled consistency index (RC), were calculated.
BLASTn search was conducted to find the genetic identity of our sequences with reference sequences in GenBank. The results for ITS showed 100% identity with Erysiphe orixae (MT645846), whereas sequences of the LSU gene were 98.8% similar to E. magnifica (MW729345) and 98.7% to E. alphitoides (MF187451). No sequence data for LSU are available for E. orixae in GenBank. Moreover, 2 bp differences in the ITS region were found between our sequences and another sequence of E. orixae from MUMH0015 (LC009907). The final dataset used in the phylogenetic analysis consisted of 18 sequences and 1,499 characters, of which 106 (7.07%) were variable and parsimony-uninformative and 101 (6.73%) were informative for parsimony analysis. In the resulting MP tree, the two newly obtained sequences of Orixa powdery mildew were clustered in a distinct branch with two Japanese sequences of E. orixae, and this branch was supported by the highest BS value (100%) (Fig. 2).
Fig. 2
A maximum parsimony tree of Erysiphe orixae was generated based on a combined dataset of the ITS and LSU sequences. Eighteen sequences were analyzed. A sequence of Erysiphe nothofagi (AB378736) was selected as an outgroup. Isolates obtained in this study are shown in bold. Bootstrap values (≥70%) are indicated for related branches. The calculated tree scores are shown in the box. ITS, internal transcribed spacer; LSU, large subunit; TL, tree length; CI, consistency index; RI, retention index; RC, rescaled consistency index.
The name E. orixae was first introduced by Braun [10], and its detailed morphological characteristics were described and illustrated by Nomura [11]. The conidial state of the Korean samples was consistent with Nomura’s description. However, Nomura provided the foot-cells of conidiophores as 16-45 μm long, which is longer than (12-)18-24 μm found in the current study. While some conidiophores in the Korean samples exhibited long foot-cells (Fig. 1G), these long foot-cells were found to be associated with abnormal or incomplete formation of the septum, resulting in the absence of the septum at that position. Therefore, the relatively short foot-cells of conidiophores could be a good way to recognize this species.
This powdery mildew species is endemic to Japan [6,12]. In addition to E. orixae, nine fungal species have been reported on O. japonica in China and Japan, which include Caponodium salicinum, Clonostachys oblongispora, Diaporthe caryae, D. eres, D. orixae, Guignardia endophyllicola, Microsphaera alni, and M. orixae [12]. This is the first report of E. orixae powdery mildew and the occurrence of powdery mildew disease on O. japonica in Korea. This finding could help identify E. orixae in its conidial state and offer valuable insights into powdery mildew diversity in Korea.


