INTRODUCTION
The genus Helvella (Helvellacae, Peziziomycetes) is characterized by saddle or cup-shaped fruiting bodies with lobes or wrinkles, and is commonly found in valleys, mossy areas, or grassy fields. Some Helvella species play an important ecological role in forest ecosystems by growing mycorrhizae [1], while H. jocatoi and H. bachu are used as food ingredients in some countries [2,3], Therefore, Helvella species are valuable forest resources.
Research on the Helvella genus has previously been conducted in Europe and the USA, with a primary focus on species diversity and phylogenetic relationships [4-13]. In Europe, using morphological characteristics and molecular biology data, the boundaries of some challenging-to-distinguish Helvella species have been clearly delineated, revealing that many Helvella species exhibit regional endemism [8]. This highlights the need for future research on Asian Helvella species. Since then, active research on Asian Helvella species has been conducted in China, revealing the presence of numerous cryptic species within Helvella and highlighting the differences in their distribution between Europe and Asia [9,11,13-15]. New and unrecorded Helvella species continue to be reported worldwide. In Korea, there had been no reports of H. costifera prior to its discovery in 2010 [16].
In this study, Helvella spp. discovered in Taebaek City were analyzed using morphological and molecular biological approaches. In the present study, we confirmed that this species has not been recorded in Korea; therefore, it was listed as an unrecorded species.
MATERIAL AND METHODS
Morphological observation
A fungal diversity survey was conducted between July and August, 2023 in Taebaek City, Korea. Before collection, sample photographs were taken using a camera (Olympus OM-D E-M1 Mark III, Tokyo, Japan), and GPS coordinates and habitat information were recorded. The collected samples were air-dried at 40℃ for 72 h for the purpose of DNA extraction. Microscopic observations were performed using an optical microscope (BX53, Olympus, Tokyo, Japan). Microstructural measurements were performed using a camera (DP22, Olympus, Tokyo, Japan) integrated with a microscope and its software (DP2-SAL, Olympus, Tokyo, Japan). Twenty ascospores and ten microstructures, excluding the ascospores, were observed.
DNA extraction and phylogenetic analysis
For the purpose of phylogenetic analysis, genomic DNA was extracted from the specimens using Exgene Plant SV Mini (Geneall, Seoul, Korea). Polymerase chain reaction (PCR) was performed to amplify partial sequences from two partial genes (internal transcribed spacer [ITS] and nuclear ribosomal large subunit [nrLSU]) using previously published primer sets. The primer sets used in this study were: ITS1 and ITS4 for ITS [17], and hLSUf and hLSUr for nrLSU [18] (Table 1). Amplification reactions were performed in 20 μL reaction volumes containing 1 μL DNA template, 1 μL per primer (10 μM), 10 μL GainBlue™ Hot Start Master Mix, 2X (GainBio, Daejeon, Korea), and 7 μL ddH2 O. The PCR conditions involved an initial pre-denaturation step at 95℃ for 4 minutes, followed by 40 cycles of denaturation at 95℃ for 30 seconds, annealing at 64℃ for 30 seconds, extension at 72℃ for 60 seconds, followed by a final extension step at 72℃ for 10 minutes. PCR products were visualized on a 1% agarose gel and purified using an Expin PCR SV kit (GeneAll, Seoul, Korea). Sanger sequencing was performed by Cosmogenentech (Daejeon, Korea). Sequences were aligned and edited in MUSCLE 5.1 [19] using default parameters. Ambiguously aligned regions and gaps in the alignment were excluded before analysis. Alignments were constructed separately for each gene fragment using MUSCLE 5.1 and then concatenated using Geneious Prime 2023.2 (https:// www.geneious.com). Poorly aligned sites were identified using Gblocks v. 0.91b [20] with the default parameters. All the identified ambiguous sites were excluded from the analysis. All phylogenetic analyses were performed based on maximum likelihood (ML) analysis in IQtree2.2.2.7 [21] available on the CIPRES Web portal [22] using a TIM+I+G model with 1,000 bootstrap replicates. The optimal substitution model was selected using jModelTest 2.1.6 [23].
RESULTS
Molecular phylogenetics
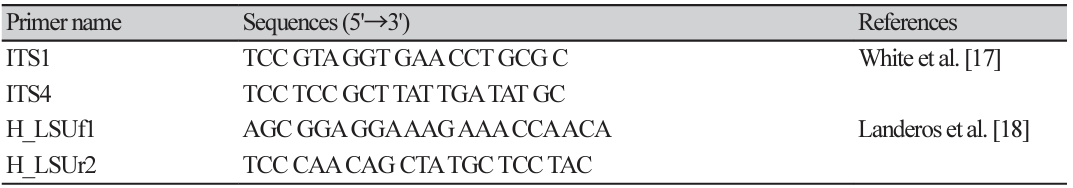
The nucleotide sequences of the 31 samples (a combination of from the ITS and nrLSU genes) were used to construct a phylogenetic tree. Reference sequences for this analysis were obtained from Zhao's study [14]. The taxa included in the phylogenetic tree comprised five species belonging to the Crispa clade (H. crispa, H. orienticrispa, H. involuta, H. pseudoreflexa, H. zhongtiaoensis) and one species belonging to the Lacunosa clade (H. rugosa). Except for H. crispa, all samples were collected from China [14]. The nucleotide sequence of the sample (KH058) used in this study, which was collected from the southwestern region of China, exhibited high bootstrap support (96%) and formed a monophyletic clade. In particular, it demonstrated a monophyletic relationship with the reference specimen H. orienticrispa (HKAS74319) with 99% bootstrap support (Fig. 1).
Fig. 1
Phylogenetic tree inferred from maximum likelihood (ML) analysis using ITS-nrLSU sequence. Sequence generated for this study are highlighted in blue. Type species are indicated in bold. ITS, internal transcribed spacer; nrLSU, nuclear ribosomal large subunit; SW, Southwestern China; NE, Northeastern China.
Taxonomy
동양주름안장버섯(신칭) Helvella orienticrispa Q. Zhao, Zhu L. Yang & K.D. Hyde, in Zhao, Tolgor, Zhao, Yang & Hyde, Phytotaxa 239(2): 136 (2015)
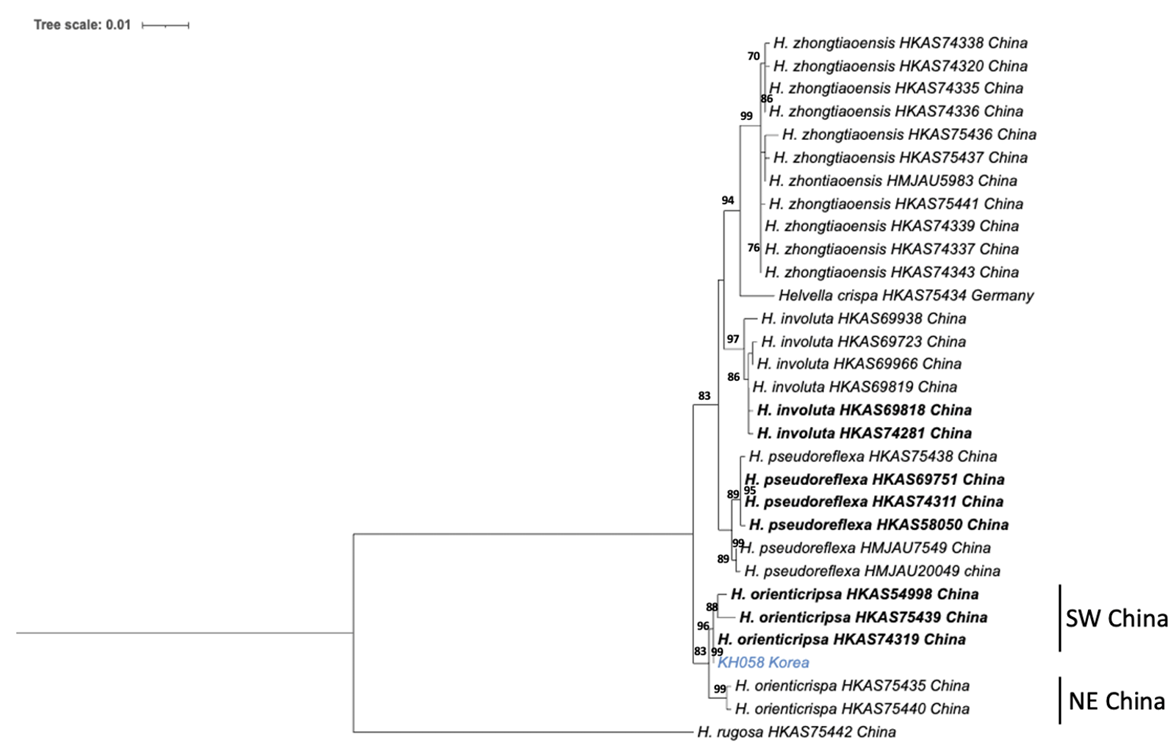
Apothecium broadly saddle-shaped to irregularly lobed, 3-4 cm high, 2-4 cm broad; margin rolled to hymenium when young, but extended and split at maturity, never fused with stipe; hymenium glabrous, pale cream to white, becoming yellowish when dried; receptacle surface finely pubescent, blunt ribs covering almost 1/3 of apothecium, pale cream to white, and yellowish when dried. Stipe 3-7 cm long, 1-1.5 cm thick, with deep lengthwise grooves and cross-veins; ribs blunt, mostly double-edged, finely pubescent, slightly brittle, pale cream to white, cream when dried, basal mycelium white. Medullary excipulum 130 -260 μm broad, textura intricata, hyaline, composed of thick-walled hyphae 2.5-6 μm broad. Ectal excipulum 70-200 μm, outmost cells catenuliform in long fascicled tufts, hyaline, evenly indigo blue in cotton blue with cylindrical to subclavate, slightly thick-walled end cells 24-36×10-14 μm. Asci 264312×10-18 μm, 8-spored, subcylindrical to clavate. Ascospores (16.2-) 18.3-18.5 (-19.8)×(10.6-) 12.112.3 (-13.5) μm, [Q=(1.38) 1.51-1.53 (1.73) (Qav =1.52±0.09)], broadly ellipsoid to ellipsoid, walls smooth, slightly thickened. Paraphyses filiform, 3-4.2 μm broad, slightly exceeding the asci, with a yellow refractive content in Cotton blue; apex slightly enlarged 6-8.3 μm broad (Fig. 2).
Fig. 2
Morphological characteristics of Helvella orienticrispa. A-C: Ascomata of KH058. D: Asci of KH058 in water. E: Ascospores of KH058 in water. F: Ectal excipulum of KH058 in cotton blue. (Scale bar: A-C=5 cm; D, F=50 µm; E=20 µm).
Habitat.
Solitary or scattered on the ground, in Larix kaempferi (Lamb.) Carrière and Quercus mongolica Fisch. ex Ledeb. forest
Specimen examined. Republic of Korea, Taebaek City, Geomryongso, Gangwon Province, 22 Jul. 2023 (KH058); N 37.23058. E 128.03514. GenBank no. OR647322 (ITS), and OR647324 (nrLSU).
Remark.
H. orienticrispa is very similar to the species recorded in Korea namely, H. crispa, to the extent that it is difficult to distinguish between them with the naked eye. However, H. crispa typically has irregularly shaped apothecium, with fine brown hairs on the receptacle surface [24], while H. orienticrispa is characterized by typically having broad saddle-shaped ascocarps and fine white tufts on the receptacle surface, allowing for differentiation.
DISCUSSION
H. orienticrispa, a previously unrecorded species in Korea, was discovered in the present study, and grows mainly in coniferous forests [14]. This species is distinguished from other Helvella species by its broadly spread, saddle-shaped apothecium and its upper and outer surfaces of the ascocarp cup, which are both cream colored and have fine white hairs on the outer surface. On the other hand, H. crispa, was f irst reported in Europe in 1823; as phylogenetic classification studies of Helvella species have progressed worldwide, it has been found to possess regional endemism, and several cryptic species with very similar morphologies have also been identified [8,11,14]. Recent phylogenetic classification studies of Helvella species in China suggest that H. crispa is not present in China [11], instead, several new species similar to H. Crispa, such as H. involuta, H. orienticrispa, H. pseudoreflexa, and H. zhongtiaoensis, have been recently reported [14]. Therefore, it is necessary to evaluate the presence or absence of H. crispa in South Korea.