INTRODUCTION
Since long time, apples and pears have been in the spotlight as internationally important crops to the extent that more than 100 million tons of apples and pears are produced worldwide annually [1-3]. In 2020, Korea produced approximately 550, 000 tons of apples and pears. Soil is indispensable for growing apple and pear trees. Soil supports the roots of trees and stores moisture; various interactions, such as helping or hindering the growth of trees by various rhizosphere microorganisms, occur in the soil [4,5].
Diverse fungi infect apple and pear trees [6]. Orchard soil serves as a reservoir for these pathogenic fungi, as they can overwinter in soil. We investigated fungi from apple and pear trees and orchard soils in diverse places to monitor diseases. Diverse fungi, including pathogenic fungi, were identified. Among the identified fungi, seven species belonging to the phylum Ascomycota have not been recorded in Korea. In this paper, we report the full descriptions and provide illustrations of their morphological characteristics and molecular phylogenetic positions.
MATERIALS AND METHODS
Collecting rhizosphere soil samples: The rhizosphere soils of apple and pear trees were collected from orchards in Cheongju, Anseong, and Cheonan, Korea. After excavating approximately 30 cm from the surface, the soil (approximately 100 g) around the roots of apple and pear trees was collected using a spade and collected in a 50 mL conical tube. Soil-containing tubes were stored in a cooler and transported to the laboratory for fungal isolation.
Isolation of fungi: Further, 3 g of the collected soil was placed in a 50 mL conical tube containing 10 mL of sterile distilled water and vortexed for 30 min. The mixture was then diluted up to 1,000 times through serial dilution; 1 mL of muddy water was mixed with 9 mL of sterile distilled water. Diluted muddy water (100 µL) was dispensed onto Dichloran-Glycerol 18% (DG18) agar medium and spread using a glass spreader. DG18 agar plates spread with soil dilution were incubated for at least three weeks in an incubator at 25℃. Among the growing fungal mycelia, mycelia that appeared different in morphology were collected and separated in pure form on potato dextrose agar (PDA). Purely isolated strains were cultured on PDA, malt extract agar (MEA), czapek yeast extract agar (CYA), and oatmeal agar (OA) to observe colony morphology.
Microstructure observation: The microstructures of the isolated fungi were observed using an optical microscope (BX53; Olympus, Tokyo, Japan). The size of each microstructure was measured 50 times, with the range shown.
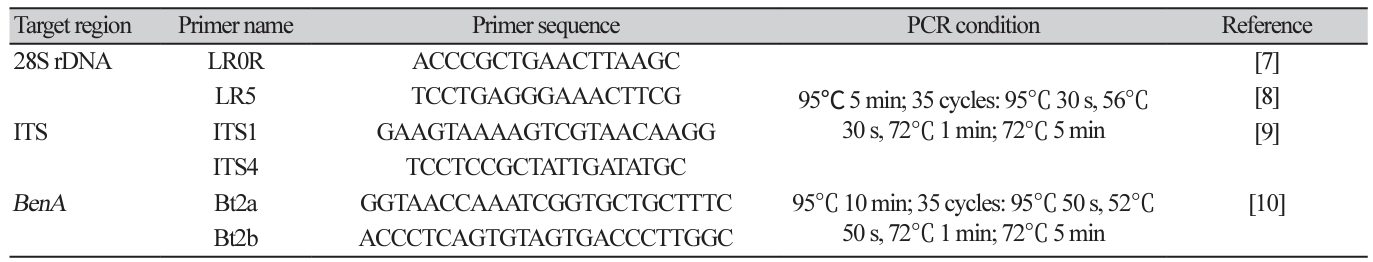
DNA extraction and phylogenetic analysis: Purely isolated fungi were cultured on PDA lined with sterilized cellophane for another week and approximately 50 mg of mycelia was scraped with a surgical blade and collected in a 2 mL tube. DNA was extracted from the collected mycelia using a Plant/Fungi DNA Isolation Kit (Norgen Biotek Corp., Thorold, Canada). Using the extracted DNA as a template, PCR primers in Table 1 were used to amplify the internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2), the large subunit of nuclear ribosomal RNA (28S rDNA), and partial β-tubulin gene (BenA) sequences. PCR was performed under the conditions listed in Table 1 using a Bio-Rad T100 Thermal cycler. After confirming PCR product amplification by electrophoresis on a 0.8% agarose gel, the PCR amplicon was purified and sequenced by Macrogen Corp. (Seoul, Korea).
The determined nucleotide sequences were analyzed for homology using the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) search engine GenBank BLASTN. Fungal species closely related to the taxa isolated in this study and belonging to the same or different genera, were included in the analysis. Reference sequences were obtained from GenBank and are listed in Tables 2-8. Individual sequence datasets of the ITS region, 28S rDNA, and the β-tubulin gene were aligned using the MUSCLE (multiple Sequence Comparison by Log- Expectation) alignment tool of MEGA XI [11] and improved manually where necessary using BioEdit [12]. Based on the aligned sequences, a maximum likelihood (ML) phylogenetic tree was constructed and the reliability of each node in the phylogenetic tree was evaluated using 1,000 bootstraps [13].
RESULTS AND DISCUSSION
The morphological characteristics of each species of unrecorded soil fungi isolated and identified in this study and the results of phylogenetic analysis based on nucleotide sequence analysis of ITS and 28S rDNA are presented below. The seven DUCC strains were deposited at the National Institute of Biological Resources of the Republic of Korea (https://www.nibr.go.kr/cmn/main/enMain.do) and received the NIBRFGC numbers. The analyzed nucleotide sequences were registered in GenBank of the NCBI database and the registration numbers of the seven DUCC strains are provided in Tables 2-8.
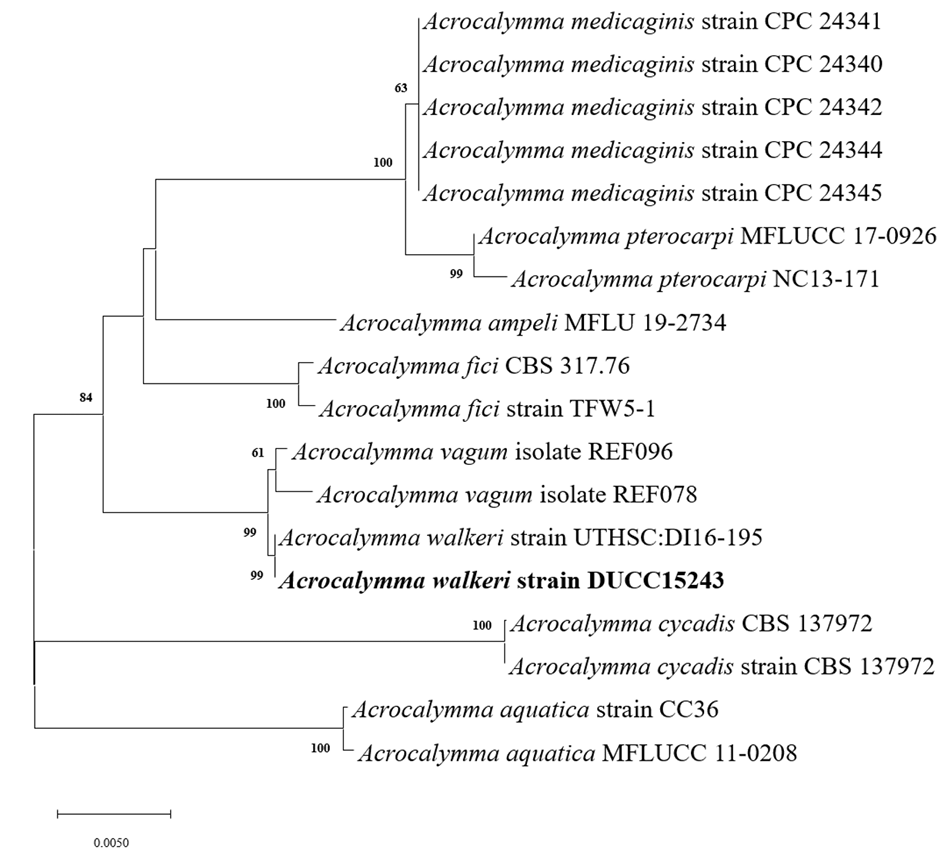
Acrocalymma walkeri Trakunyingcharoen et al. IMA Fungus 5(2):391-414 (2014) [MB#810840]
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Acrocalymma walkeri DUCC15243 (NIBRFGC000509977) grew the best with a diameter of 38-40 mm on MEA and CYA and grew the slowest with a diameter of 26-31 mm on OA. The colonies were generally white, slightly fluffy, and circular and had few developed aerial hyphae. However, DUCC15243 showed a light pink colony color on OA. Microscopic morphological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and 2-3 μm thick. Conidia were hyaline, with a smooth surface, without septum, olive colored, oval, filled with oily droplets, and 14.5-16.2×3.2-4.1 μm thick (Fig. 1). No sexual state was observed. A. walkeri was isolated from Medicago sativa in Queensland, Australia by Crous and Trakunyingcharoen in 2014 [14]. A. walkeri has been reported to have a similar morph whereas larger conidia (11-21×3.5-5 µm) compared to those of A. medicaginis. Molecular biological characteristics: The ITS region of the DUCC15243 strain showed 99.79% similarity with that of A. walkeri LT796832 and the 28S rDNA region showed 99.55% similarity with that of A. walkeri MH874057. According to the phylogenetic tree based on the combined ITS and 28S rDNA sequences shown in Table 2, DUCC15243 strain formed the same clade as A. walkeri strain UTHSC:DI16-195 (Fig. 2). Based on the morphological and molecular analysis, DUCC15243 strain was identified as Acrocalymma walkeri, an unrecorded species in Korea. A. walkeri causes root and crown rot in Medicago sativa [15]. A. vagum belonging to the same genus as A. walkeri produces dibenzo-α-pyrone derivatives with antibacterial and antifungal activity [16] and licorice, a medicinal plant, has been reported to tolerate drought stress better when its rhizosphere was treated with A. vagum [17]. Therefore, it would be interesting to study whether A. walkeri can produce substances with antibacterial and antifungal activities and interact with plants in the rhizosphere.
Fig. 1
Colony morphology on different media (A-D) and light microscopic images (E-H) of Acrocalymma walkeri DUCC15243 (NIBRFGC000509977) strain grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. E, F: Conidiogenous cells. G, H: Conidia. Scale bar=10 μm.
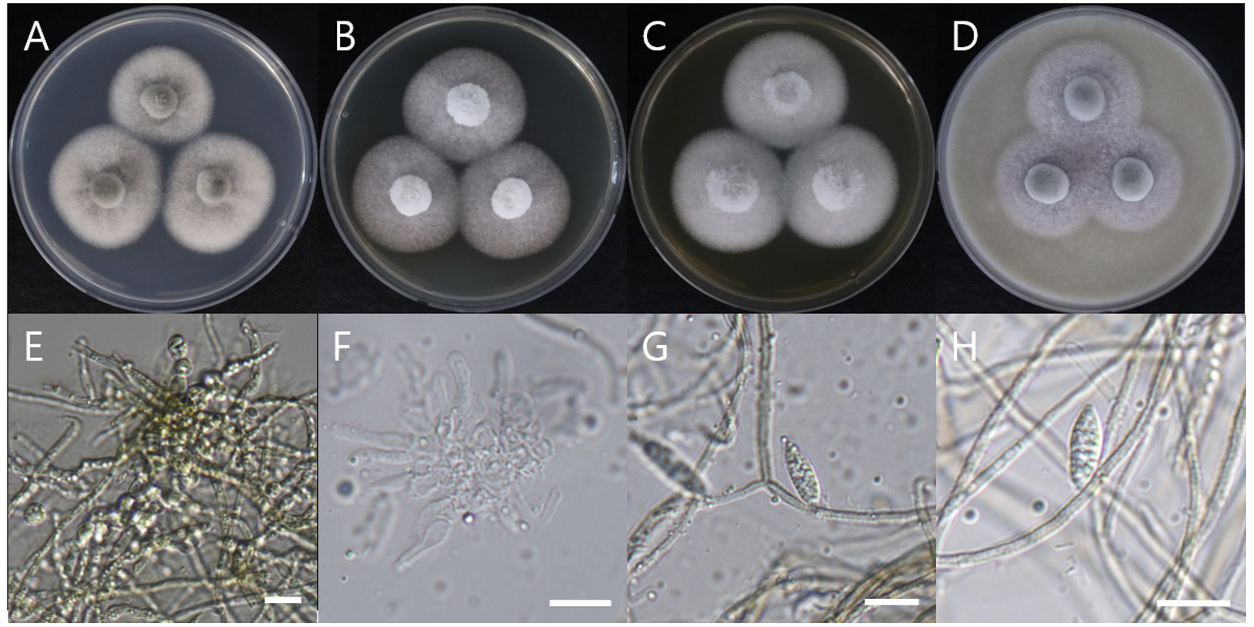
Table 2
Sequence information of species closely related to Acrocalymma walkeri used for constructing a phylogenetic tree based on the concatenated sequences of ITS and 28S rDNA region
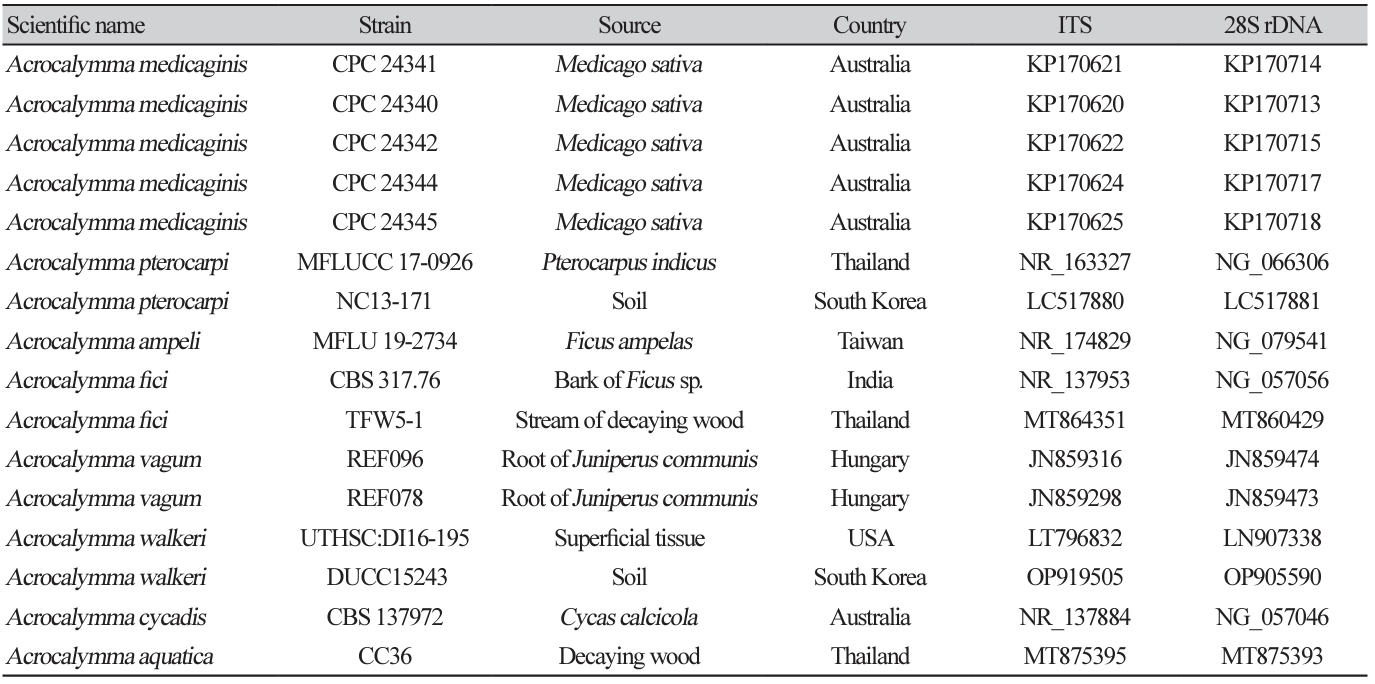
Fig. 2
Maximum likelihood phylogenetic tree based on the concatenated ITS region and 28S rDNA nucleotide sequences of Acrocalymma walkeri DUCC15243 (NIBRFGC000509977) strain. The number of nodes represents the reliability value through 1,000 bootstrap replicates. The fungal strain isolated in this study is indicated in bold.
Clonostachys krabiensis Tibpromma et al. Fungal Diversity 93:1-160 (2018) [MB#554527]
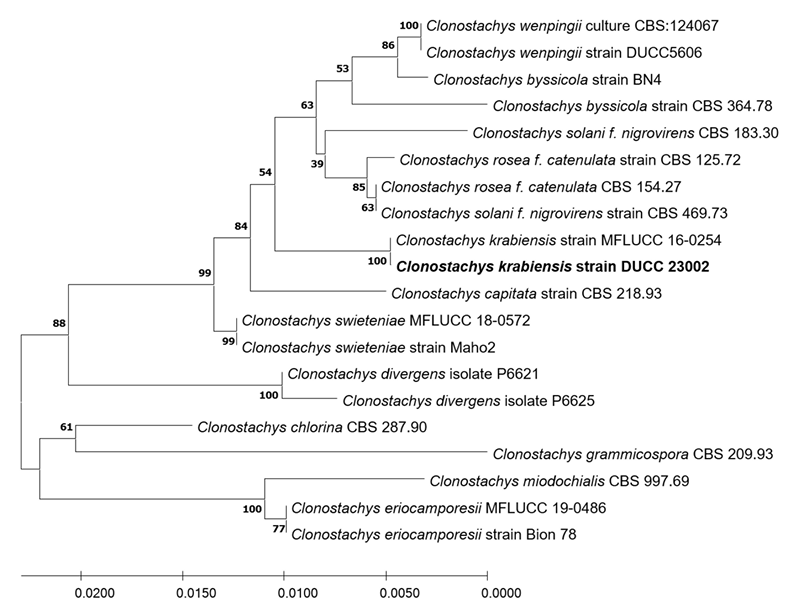
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Clonostachys krabiensis DUCC23002 (NIBRFGC000510016) were fluffy, circular, radial, and white and developed aerial hyphal morphologies on all media. The DUCC23002 strain grew best, with a diameter of 34 mm on PDA and showed the slowest growth on OA. Microscopic morphological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and 1-3 μm thick. Conidiophores were hyaline and thin-walled; developed at the tip or middle of the hyphae with a size of 13.7-25.1×2.6-4.6 μm. Phialides were hyaline and thin-walled; 1-3 developed at the end of the conidiophore, with a size of 14.2-30.1× 2.3-4.5 μm. Conidia had a smooth surface, no septum, olive color, oval shape, and a size of 3.4-5.6×2.55.0 μm (Fig. 3). No sexual state was observed. C. krabiensis was isolated from Pandanus spp. in Thailand by Tibromma in 2018 [18]. C. krabiensis has been reported to have conidiophores that aggregate into sporodochia with hyaline setae around the margin. The conidiogenous cells were monophialidic, hyaline, subulate, and 10-13×1.5-2.5 μm in size. Conidia were slimy, solitary, aseptate, hyaline, cylindrical to oblong, smooth-walled, and 5-7×1-2 μm in size. Molecular biological characteristics: The ITS region of the DUCC23002 strain showed 98.49% similarity with that of C. krabiensis NR_168189 and the 28S rDNA showed 99.76% similarity with that of C. krabiensis NG_068835. According to the phylogenetic tree based on the combined ITS region and 28S rDNA sequences in Table 3, DUCC23002 strain formed the same clade as C. krabiensis strain MFLUCC 16-0254 (Fig. 4). Based on morphological and molecular analysis, DUCC23002 strain was identified as Clonostachys krabiensis, an unrecorded species in Korea. C. rhizophaga belonging to the genus Clonostachys is a plant pathogen that causes chickpea wilt [19]. As a fungal parasite, C. rosea has strong biological control abilities against numerous fungal plant pathogens, nematodes, and insects [20]. However, scarce information is available on C. krabiensis and additional research should be conducted to determine its value and role in nature.
Fig. 3
Colony morphology on different media (A–D) and light microscopic images (E-H) of Clonostachys krabiensis DUCC23002 (NIBRFGC000510016) strain grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. E-G: Conidiophore. H: Conidia. Scale bar=10 μm.
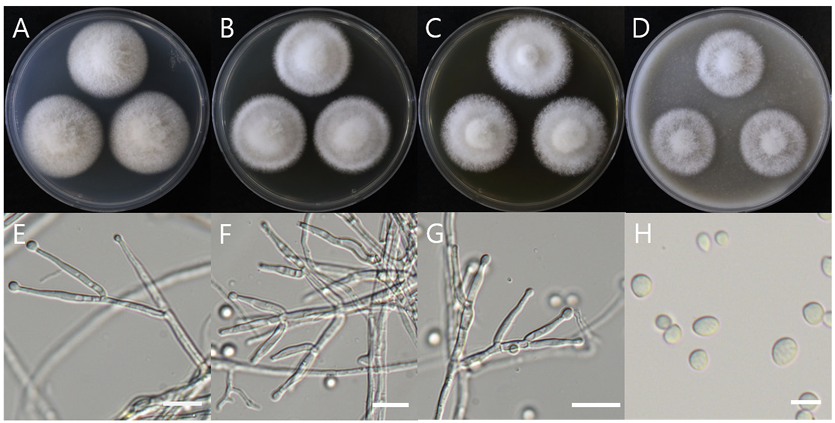
Table 3
Sequence information of species closely related to Clonostachys krabiensis used for constructing a phylogenetic tree based on the concatenated sequence of ITS and 28S rDNA region
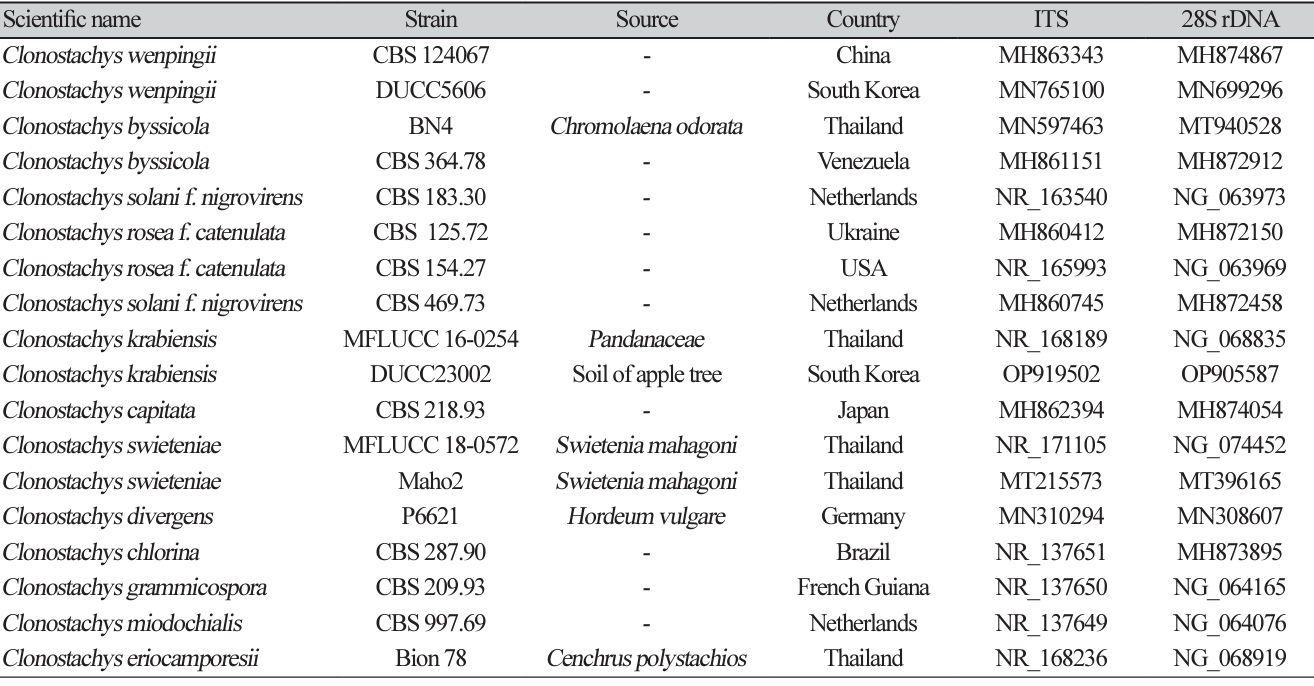
Fig. 4
Maximum likelihood phylogenetic tree based on the concatenated ITS region and 28S rDNA nucleotide sequences of Clonostachys krabiensis DUCC23002 (NIBRFGC000510016) strain. The number of nodes represents the reliability value through 1,000 bootstrap replicates. The fungal strain isolated in this study is indicated in bold.
Coniella vitis Chethana et al. Plant Disease 101(12):2123-2136 (2017) [MB#819365]
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Coniella vitis DUCC15252 (NIBRFGC000509981) strain grew rapidly on all media and had a colony size of 80-85 mm. The colonies were white and circular, with mycelial density decreasing toward the outer periphery on PDA and OA. Irregular colony morphology was observed on MEA. On CYA, it was a circular shape with a slightly wavy pattern. Molecular biological characteristics: Hyphae were hyaline, unbranched, septate, thin-walled, and 1-5 μm thick. The pycnidia were greenish-black to the naked eye, spherical, light brown under a light microscope, thick-walled, and 73-310×47-96.8 μm thick. Conidia were smooth, without septum, olive-colored, oval, and 6.7-13.1×3.8-10.7 thick (Fig. 5). C. vitis was isolated from diseased grapevines in the Beijing, Guangxi, Hebei, Henan, and Jilin provinces of China by Chethana in 2017 [21]. However, the original description did not clearly describe the morphological characteristics of C. vitis. However, a rough comparison showed that DUCC15252 was very similar to C. vitis. Molecular biological characteristics: The ITS region of the DUCC15252 strain showed 99.82% similarity with that of C. vitis OM283102 and the 28S rDNA region showed 100% similarity with that of C. vitis MH569461. According to the phylogenetic tree based on the combined ITS and 28S rDNA sequences in Table 4, the DUCC15252 strain formed the same clade as the C. vitis strain MFLUCC 18-0093 (Fig. 6). Based on the morphological and molecular analysis, DUCC15252 strain was identified as Coniella vitis, an unrecorded species in Korea. Bacillus velezensis GSBZ09 could inhibit the mycelial growth and spore germination of C. vitis which is known as the causative agent of grape white rot [22].
Fig. 5
Colony morphology on different media (A–D) and light microscopic images (E-H) of Coniella vitis DUCC15252 (NIBRFGC000509981) strain grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. E, F: Pycnidia. G: Conidiogenous cell. H: Conidia. Scale bar=10 μm.
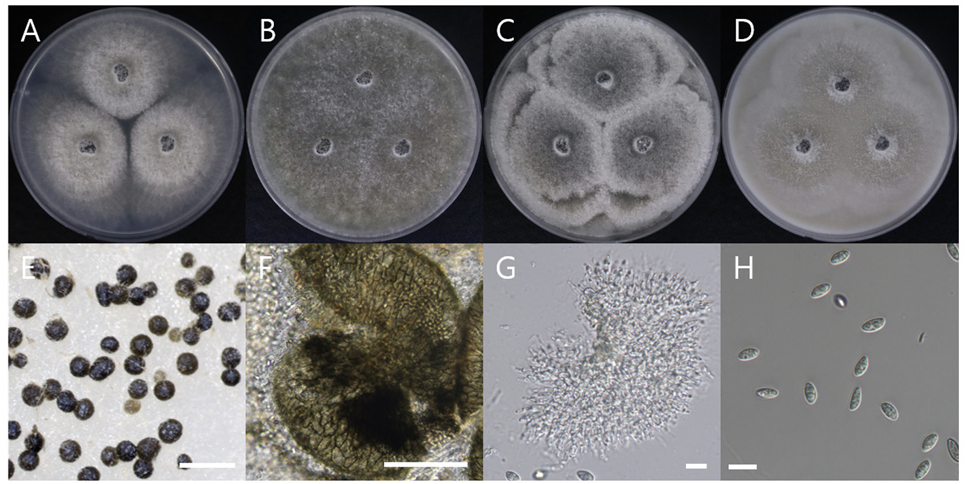
Table 4
Sequence information of species closely related to Coniella vitis used for constructing a phylogenetic tree based on the concatenated sequences of ITS and 28S rDNA region
Fig. 6
Maximum likelihood phylogenetic tree based on the concatenated ITS and 28S rDNA nucleotide sequences of Coniella vitis DUCC15252 (NIBRFGC000509981) strain. The number of nodes represents the reliability value through 1,000 bootstrap replicates. The fungal strain isolated in this study is indicated in bold.

Cosmospora diminuta Rossman et al. Studies in Mycology 42:1-248 (1999) [MB#460228]
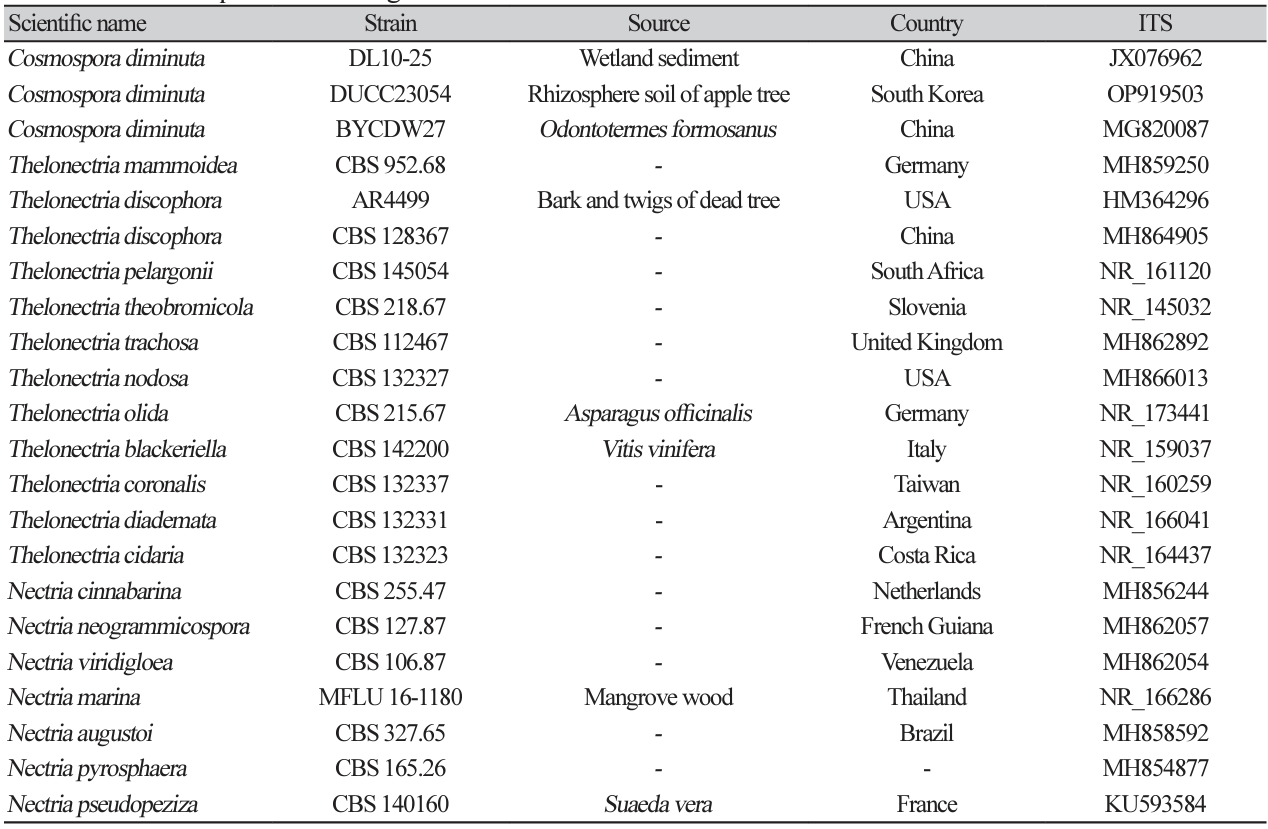
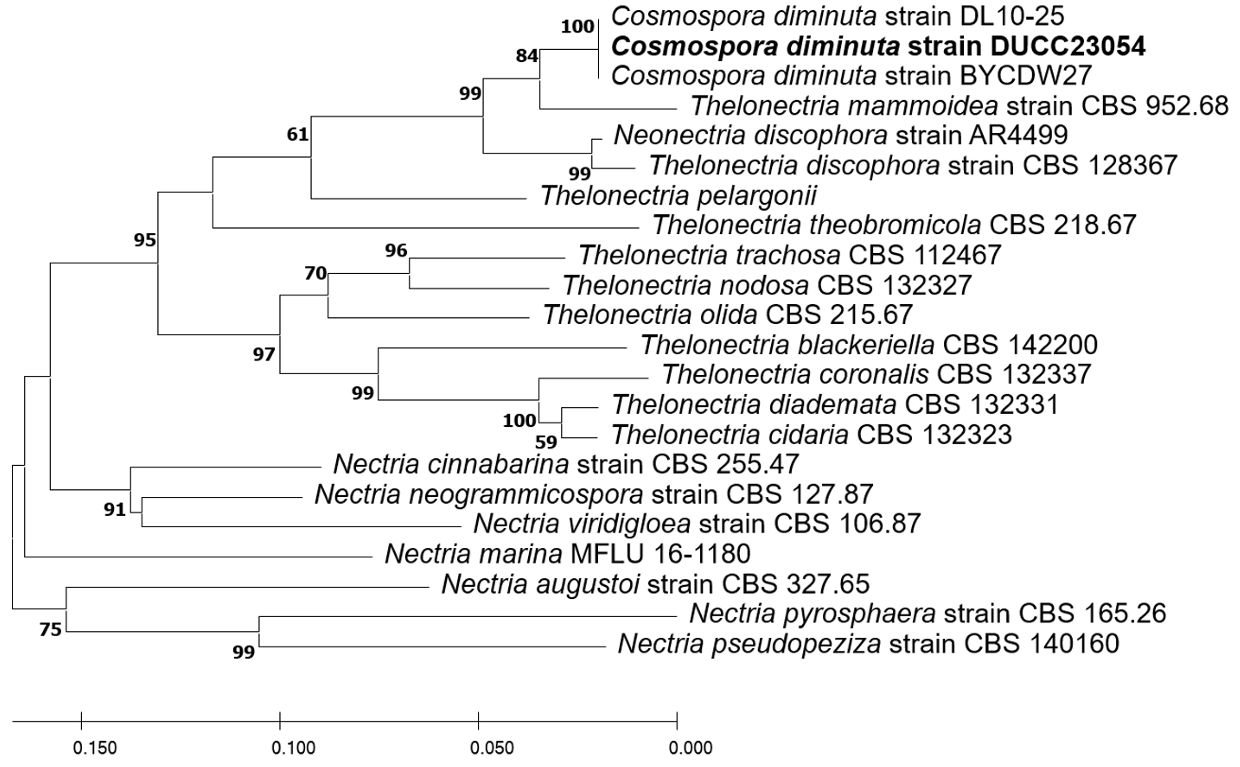
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Cosmospora diminuta DUCC23054 (NIBRFGC000510017) strain showed a colony size of 40-45 mm on PDA, OA, and MEA and grew best at 51-52 mm on CYA. The colony color was generally mixed with white and purple, and a less purple color was observed only on PDA. Overall, the colony was fluffy with slightly developed aerial hyphae and had the most developed aerial hyphae and a fluffy shape on CYA. Molecular biological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and 1.5-3 μ m thick. Conidiophores were hyaline, thin-walled, developed at the tip of the hyphae, and 15.2-37.3×2.84.5 μm thick. Phialides were hyaline, thin-walled, 1-3 developed at the end of the conidiophore, and 9.523.5×2.2-3.9 μm thick. Microconidia were smooth, without septum, oval, and 6.3-9.4×3.6-4.9 μm thick. Macroconidia were smooth, with 0-3 septum, caterpillar-shaped, found only on CYA, and 33.4-48.5×5.27.3 μm thick (Fig. 7). No sexual state was observed. Nectria diminuta was named C. diminuta comb. nov. by Rossman in 1999. In the original article, only the sexual form of C. diminuta was described; Dialonectria diminuta was expected to be an anamorph of C. diminuta [23]. However, information on D. diminuta was not sufficient to compare with that of the DUCC23054 strain as only conidiogenous cells with a size of 5-11 ×3 μm and conidia with a size of 4-5.5×3 μm were present. Molecular biological characteristics: The ITS region of DUCC23054 showed 99.27% similarity with that of C. diminuta MG820087. According to the phylogenetic tree based on the ITS sequence in Table 5, the DUCC23054 strain formed the same clade as the C. diminuta strain DL 10-25 and BYCDW27 (Fig. 8). Based on morphological and molecular analysis, the DUCC23054 strain was identified as Cosmospora diminuta, an unrecorded species in Korea. The genus Cosmospora is a mycoparasite that has been reported to have a wide range of fungal hosts [24].
Fig. 7
Colony morphology on different media (A-D) and light microscopic images (E-H) of Cosmospora diminuta DUCC23054 (NIBRFGC000510017) strain grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. E, F: Conidiophore and Phialide. G: Macroconidia. H: Microconidia. Scale bar=10 μm.
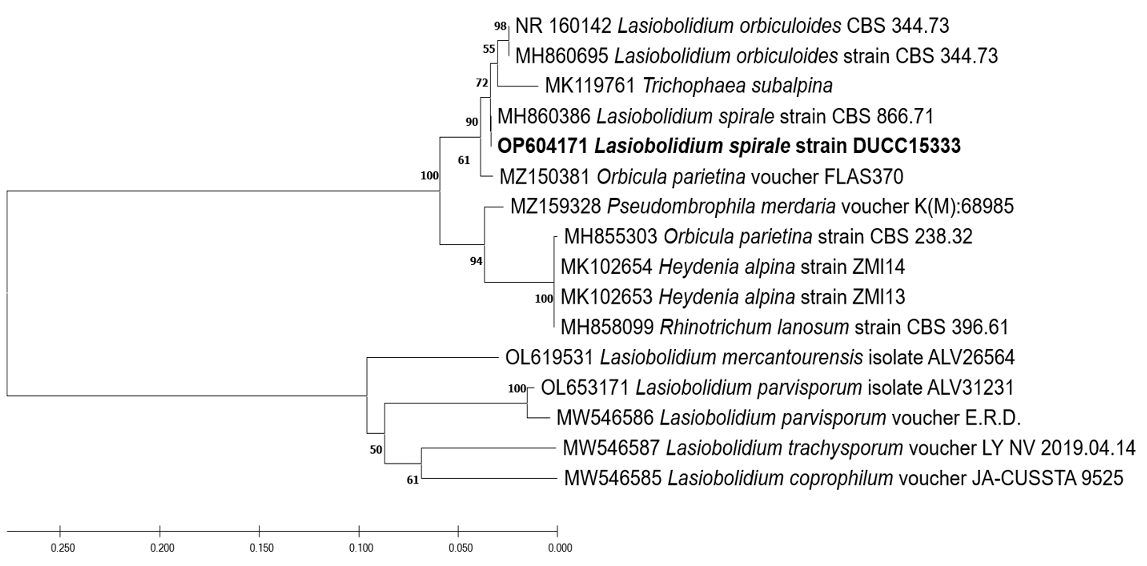
Lasiobolidium spirale Malloch and Cain. Canadian Journal of Botany 49(6):847-854 (1971) [MB#316356]
Macro morphological characteristics were observed after 5 d of incubation in the dark at 25℃. Lasiobolidium spirale DUCC15333 (NIBRFGC000510078) strain showed full growth of 90 mm on PDA and MEA whereas slightly slower growth of 67-72 mm on OA and CYA. Colonies were generally white or slightly yellowish. The shape of the colony was radial and flat on PDA, MEA, and OA, and the density of the hyphae increased toward the outside. Irregular flat shapes appeared on the CYA. Powdery microsclerotia were observed in all media. Molecular biological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and measured 1-5 μm. Microsclerotia were brown or dark brown, thickwalled, cluster-shaped, and measured 40-80×70-150 μm (Fig. 9). No sexual state was observed. L. spirale was isolated from Cryptogamic herbarium, USA, Wyoming, Albany Co., Bosler by Cain in 1964 [25]. The original description only describes the sexual morphology of L. spirale. L. spirale was described as having 170-750 μm ascocarps, 35-62×12-21 μm asci, and 12-17×9-12 μm ascospores. However, morphological comparisons were complicated as L. spirale, which we isolated from the soil, had only an asexual morphology. Molecular biological characteristics: The ITS region of the DUCC15333 strain showed 100% similarity to that of L. spirale MH860386. According to the phylogenetic tree based on the ITS region sequences in Table 6, the DUCC15333 strain formed the same clade as L. spirale strain CBS 866.71 (Fig. 10). Based on morphological and molecular analysis, DUCC15333 strain was identified as Lasiobolidium spirale, an unrecorded species in Korea, however, its other biological properties remain unknown.
Fig. 9
Colony morphology on different media (A-D) and light microscopic images (E-H) of Lasiobolidium spirale DUCC15333 (NIBRFGC000510078) strain grown at 25℃ for 5 d. A: PDA. B: MEA. C: CYA. D: OA. E-H: Microsclerotium and hyphae. Scale bar: E, F=100 μm; G, H=10 μm.
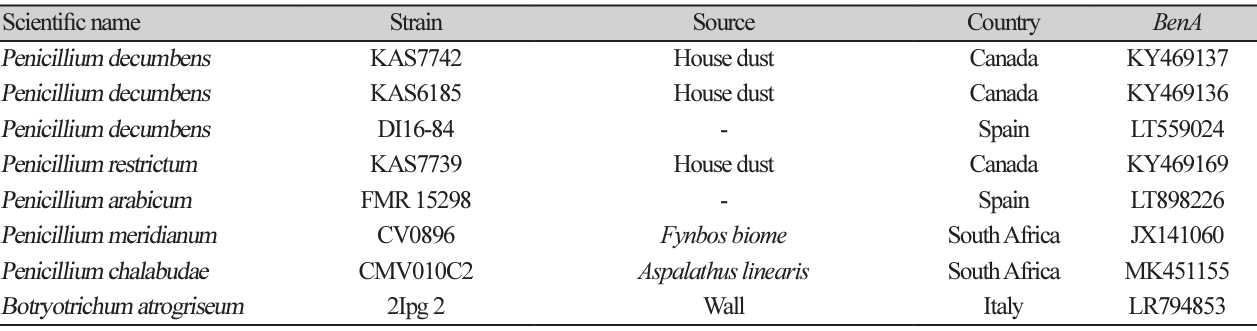
Penicillium vallebormidaense Crous et al. Persoonia 45:251-409 (2020) [MB#837659]
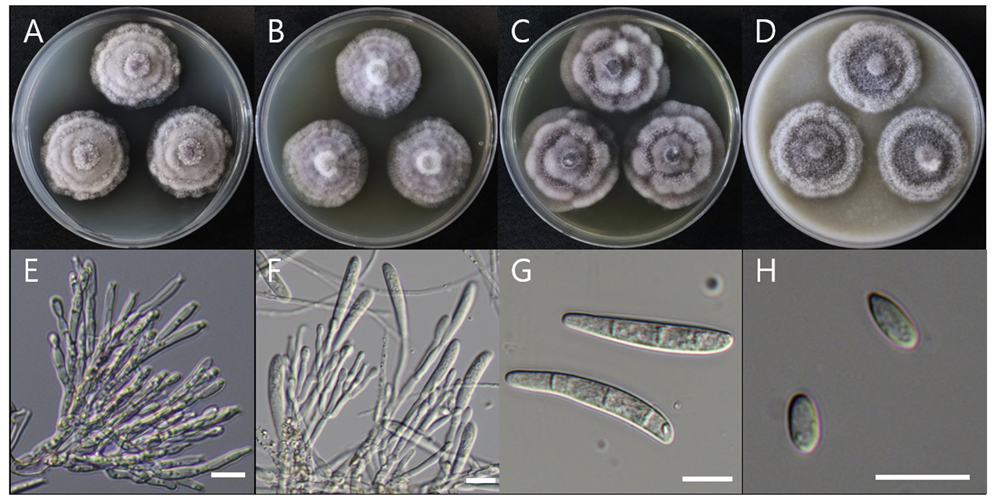
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Penicillium vallebormidaense DUCC15362 (NIBRFGC000510080) grew slowly, to 14-17 mm on CYA and MEA, and slightly faster, to 17-20 mm on OA and PDA, however, it generally grew slowly. The colony was white on CYA and MEA and yellow bands were observed on OA and PDA. The colonies were radial, circular, and slightly fluffy, with slightly developed aerial hyphae, on all media. Molecular biological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and measured 2-3 μm. Conidiophores were hyaline, without septum, thin-walled, and measured 15.6-37.2×2.2-3.0 μm. Phialides were hyaline, without a septum, thin-walled, with 2 or 3 phialides attached to matulae or conidiophore, and measured 7.3-14.7×1.7-3.7 μm. Conidia were hyaline and circular, measuring 2.4-3.5 μm (Fig. 11). No sexual state was observed. Penicillium vallebormidaense was first isolated from compost in Savona, Valle Bormida, and Ferrania in Italy by Di Piazza in 2018. In the original description, P. vallebormidaense was characterized by monoverticillate conidiophores; non-vesiculate, smooth, short stipes measuring 13-25 (-35) ×1.5-2.5 µm; and ampulliform phialides, with 3-6 per conidiophore, measuring 7-8.5 (-10)×2-2.5 (-3) µm. Conidia were smooth, globose to subglobose, and measured 2-2.5 (-3) µm [26]. These morphologies were very similar to those of the DUCC15362 strain, which was found in the rhizosphere soil of pear trees. Molecular biological characteristics: The ITS region of the DUCC15362 strain showed 100% similarity to that of P. vallebormidaense MT316359 and the BenA region of the DUCC15362 strain showed 100% similarity to that of P. vallebormidaense MW115862. According to the phylogenetic tree based on the partial BenA sequence in Table 7, the DUCC15362 strain formed the same clade as P. vallebormidaense strain DTO 402-H5 (Fig. 12). Based on morphological and molecular analysis, DUCC15362 strain was identified as P. vallebormidaense, an unrecorded species in Korea. P. vallebormidaense was relatively recently, information on its other properties is not available. However, P. menonorum, a species phylogenetically related to P. vallebormidaense, has been isolated from the rhizosphere of cucumber plants and is known to promote plant growth [27].discovered relatively recently, information on its other properties is not available. However, P. menonorum, a species phylogenetically related to P. vallebormidaense, has been isolated from the rhizosphere of cucumber plants and is known to promote plant growth [27].
Fig. 11
Colony morphology on different media (A-D) and light microscopic images (E-H) of Penicillium vallebormidaense DUCC15362 (NIBRFGC000510080) strain grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. E-G: Conidiophore and phialide. H: Conidia. Scale bar=10 μm.
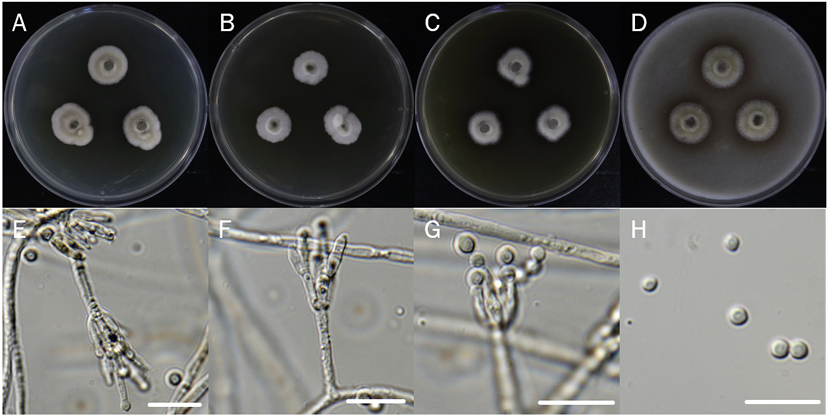
Table 7
Sequence information of species closely related to Penicillium vallebormidaense used for constructing a phylogenetic tree based on the sequence of BenA
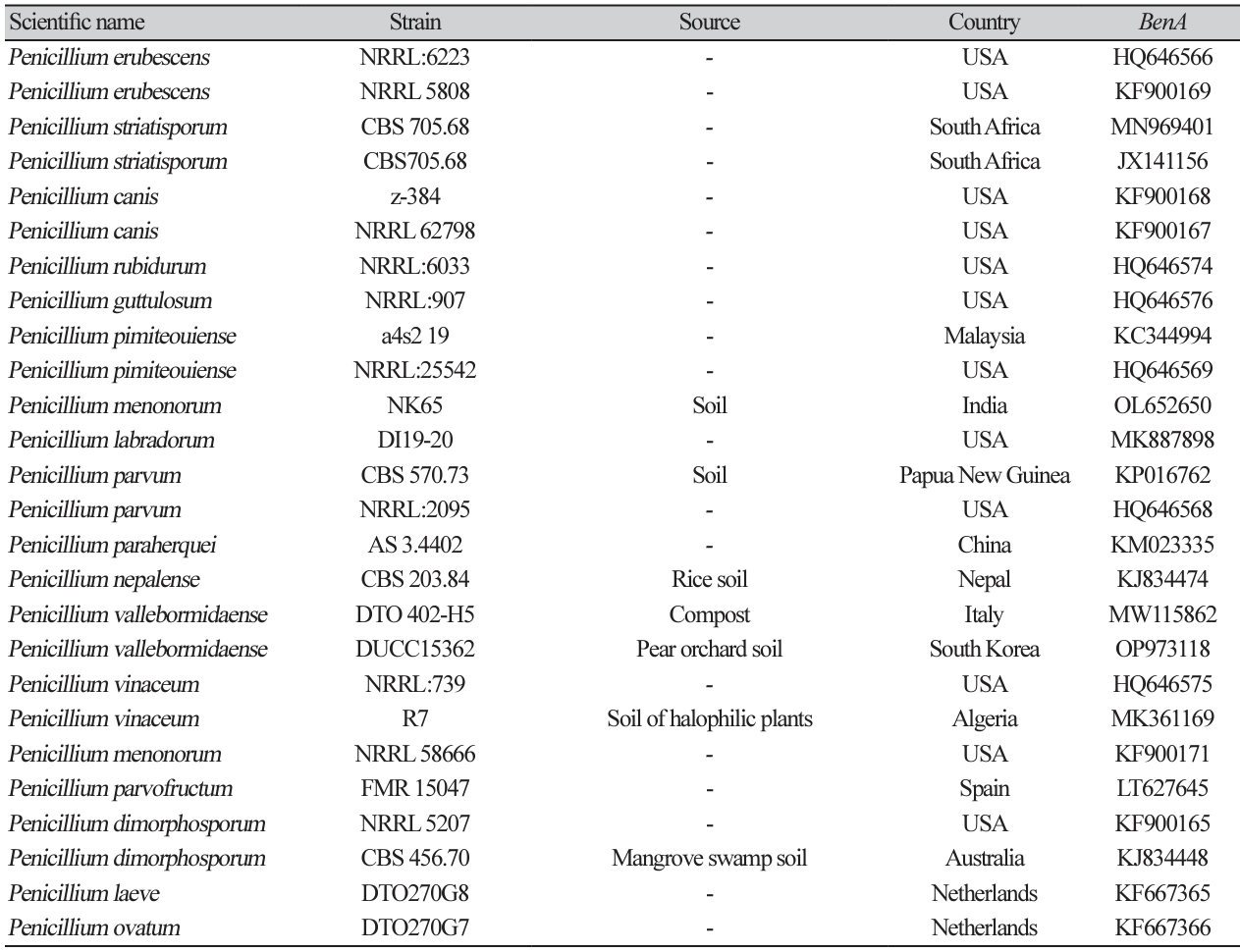
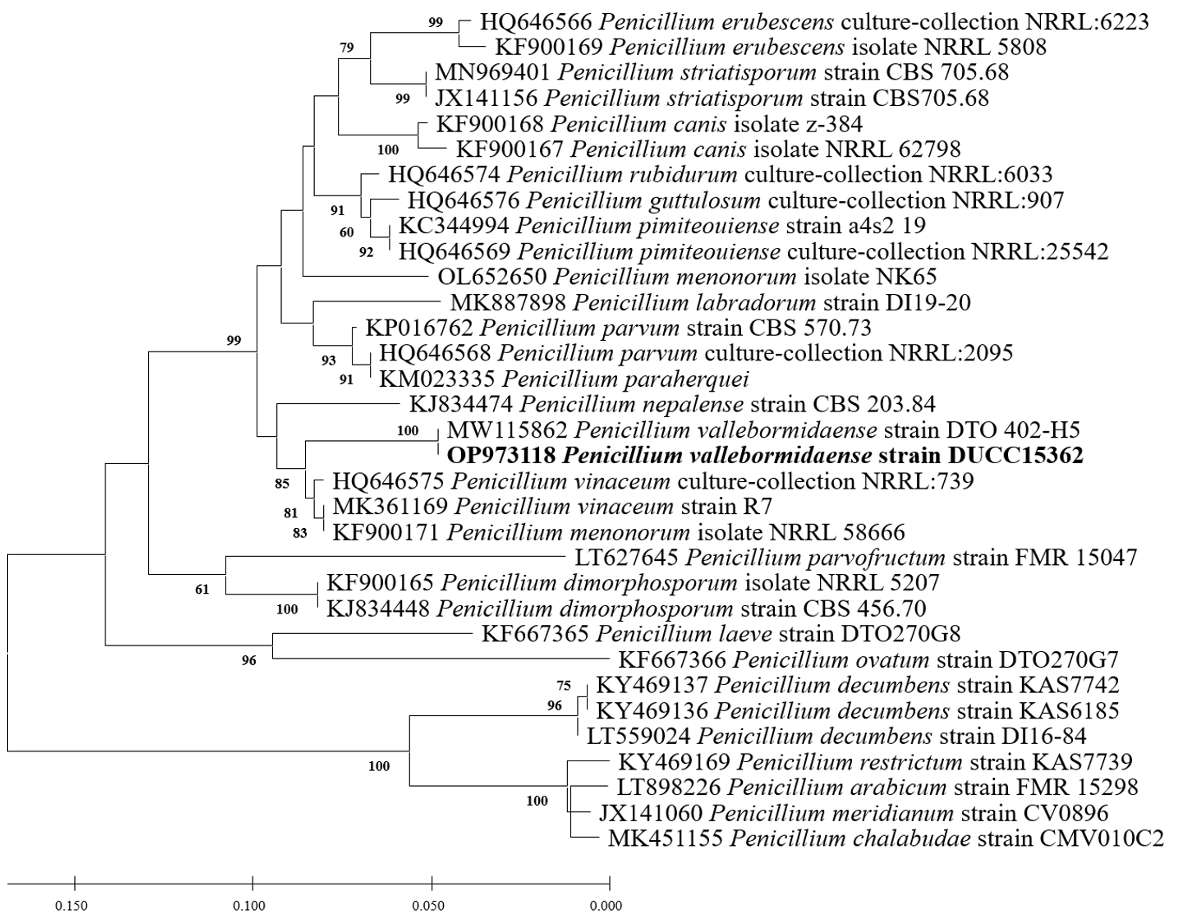
Fig. 12
Maximum likelihood phylogenetic tree based on the partial BenA sequence of Penicillium vallebormidaense DUCC15362 (NIBRFGC000510080) strain. The number of nodes represents the reliability value through 1,000 bootstrap replicates. The fungal strain isolated in this study is indicated in bold.
Pseudothielavia arxii Wang et al. Studies in Mycology 93:155-252 (2019) [MB#829873]
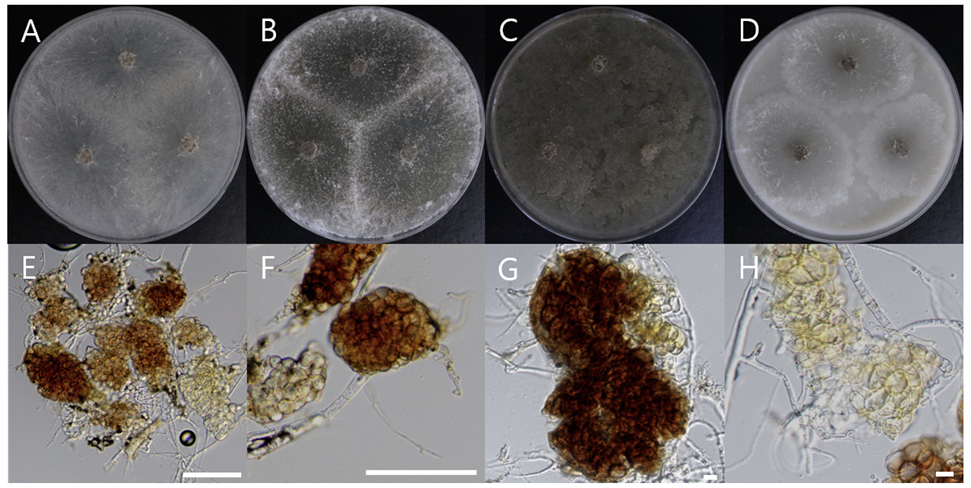
Macro morphological characteristics were observed after 7 d of incubation in the dark at 25℃. Pseudothielavia arxii DUCC22279 (NIBRFGC000510002) grew rapidly to a diameter of 76-83 mm on OA, CYA, and MEA, however, grew slowly to a diameter of 33-34 mm on PDA. The colonies were white in all media, whereas the center was gray in OA. The colony was generally round, however, lines were observed on colonies grown on PDA, though not on the rest of the medium. Overall, it was fluffy, however, was the most fluffy on the CYA medium and the aerial hyphae were also the best developed. Molecular biological characteristics: Hyphae were hyaline, branched, septate, thin-walled, and measured 2-4 μm. The ascomata were opaque, dark brown, circular, thick-walled, and measured 68.4-124.5×63.4128.5 μm. Asci were hyaline, circular, containing eight ascospores, and measured 20.6-32.8×17.5-22.5 μ m. Ascospores were hyaline, oval with pointed ends, olive green, and measured 10.5-13.8×6.4-8.2 μm (Fig. 13). P. arxii was originally isolated from the soil in Chile, Pascua Island, Hanga-Roa, by L. Zaror, date unknown [28]. In the original description, P. arxii was described as having superficial Ascomata, solitary to aggregated, non-ostiolate, and leaden black when mature in reflected light due to the dark ascomatal wall and asospores inside. The ascomata were spherical, glabrous, and measured 60-250 μm in diameter. Cells that formed the ascomatal wall were brown, semi-translucent, and densely packed. Asci were clavate to pyriform, with a spore-bearing part measuring 24-31×15-19.5 μm, and stalks 8-18.5 μm long, containing eight irregularly arranged ascospores that are evanescent. Ascospores were 1-celled, olivaceous brown when mature, smooth, fusiform, measuring (11-) 12-14.5 (-17)×(6-) 6.5-8.5 (-9.5) μm, with an oblique to lateral germ pore [28]. These morphologies were very similar to those of the DUCC22279 strain, which was found in the rhizosphere soil of pear trees. Molecular biological characteristics: The ITS region of DUCC22279 showed 99.80% similarity to that of P. arxii ON121956 and BenA of DUCC22279 showed 100% similarity to that of P. arxii MZ724098. According to the phylogenetic tree based on the partial BenA sequence in Table 8, the DUCC22279 strain formed the same clade as the P. arxii strain HGUP 190099 (Fig. 14). Based on morphological and molecular analysis, the DUCC22279 strain was identified as P. arxii, an unrecorded species in Korea. Extracts from P. arxii have been shown to inhibit the growth of Listeria monocytogenes, which causes Listeria disease [29].
Fig. 13
Colony morphology on different media (A-D) and light microscopic images (E-H) of Pseudothielavia arxii DUCC22279 (NIBRFGC000510002) grown at 25℃ for 7 d. A: PDA. B: MEA. C: CYA. D: OA. Light microscopic images; E, F: Ascomata, G: Asci and Ascospore, H: Ascospore. Scale bar=10 μm.
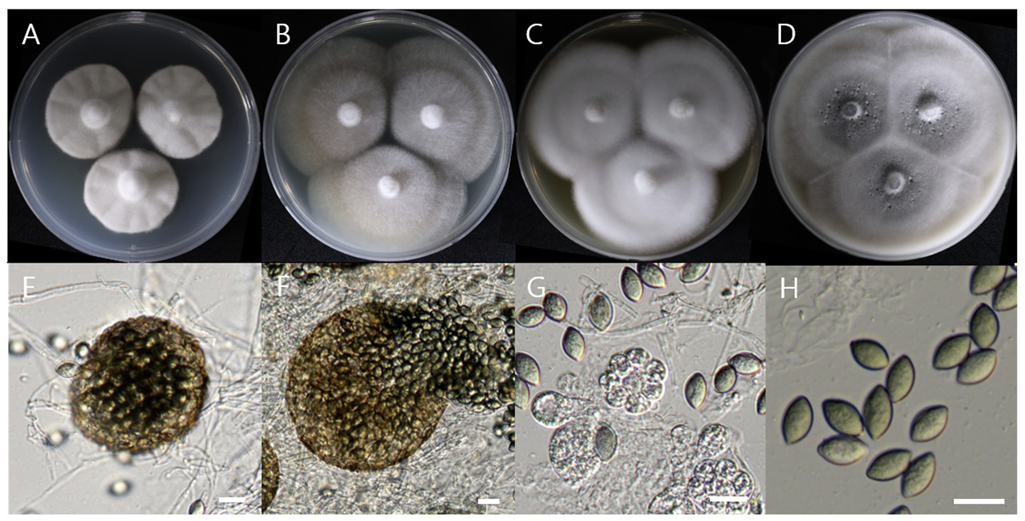
Table 8
Sequence information of species closely related to Pseudothielavia arxii used for constructing a phylogenetic tree based on the sequence of BenA
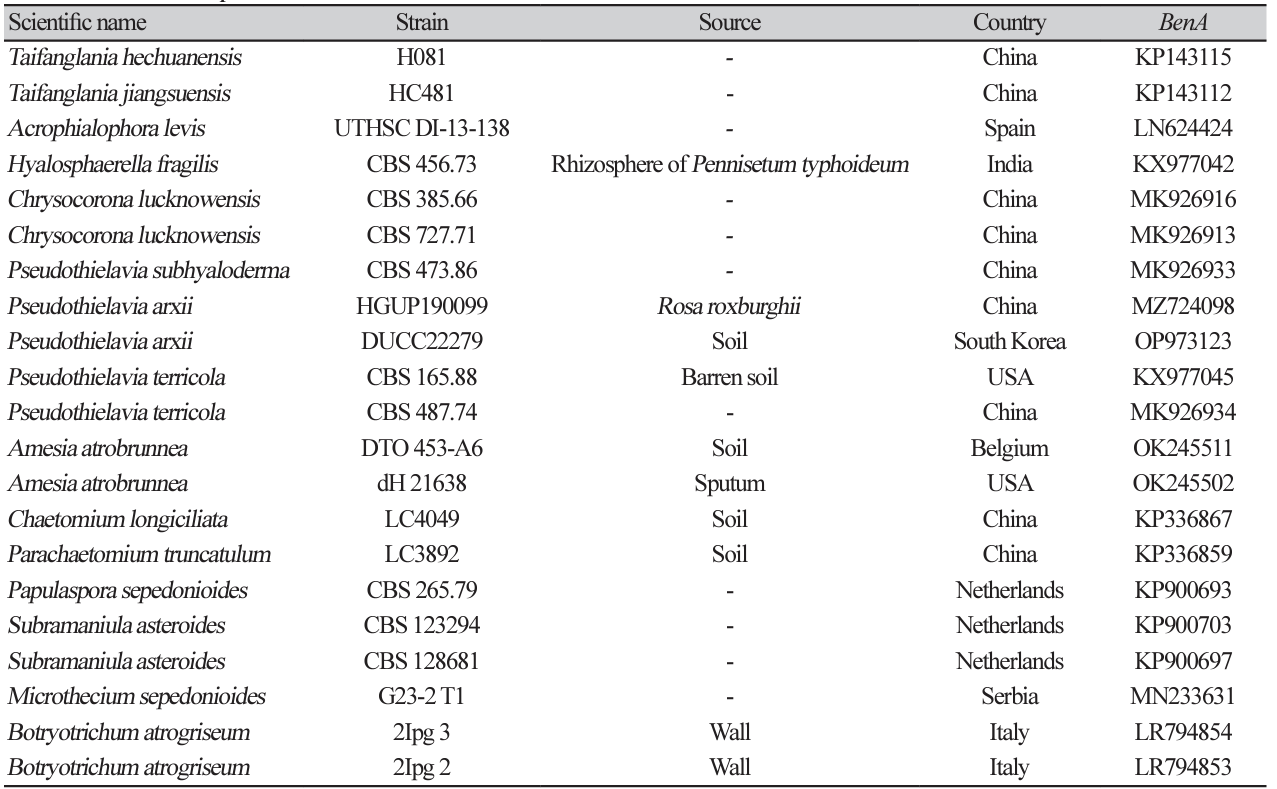
Fig. 14
Maximum likelihood phylogenetic tree based on partial BenA gene sequence of Pseudothielavia arxii DUCC22279 (NIBRFGC000510002). The number of nodes represents the reliability value through 1,000 bootstrap replicates. The fungal strain isolated in this study is indicated in bold.
Soil fungi contribute to the agricultural and horticultural ecosystem processes. They play a role in the sustainable development of agriculture [30]. They are involved in plant–soil interactions, organic matter recycling, plant growth promotion, and nutrition [31,32]. However, few studies have focused on the soil fungi in apple and pear orchards. Identification is a basic step in understanding their functions in the soil ecosystem processes in fruit tree orchards. In this study, we explored seven previously unrecorded species of Ascomycota in the rhizosphere soils of apple and pear trees in Korea. Among these seven species, A. walkeri and C. vitis are known to be plant pathogens of alfalfa and grapes in other countries. Disease outbreaks by these two pathogenic species are possible. Therefore, the presence of these two host plants in domestic ecosystems must be carefully investigated. P. arxii is a useful resource because its extracts inhibit the growth of the foodborne pathogen Listeria monocytogenes. Research on the application of this species for protecting food from foodborne pathogens is encouraged. However, information on C. diminuta, C. krabiensis, L. spirale, and P. vallebormidaense, is limited. To promote sustainable and regenerative horticulture using soil fungi, information on the biological properties of these four fungal species needs to be generated through an extended study of soil fungi in apple and pear orchards.