INTRODUCTION
Sordariomycetes is one of the largest classes of Ascomycota comprising 45 orders, 167 families, and 1,499 genera with a cosmopolitan distribution, and whose classification has changed considerably over the past decade [1-3]. The order Hypocreales, which belongs to the class Sordariomycetes, comprises 14 families, including Nectriaceae. This family includes endophytic, saprophytic, and pathogenic fungi observed in a wide range of regions [3]. The genus Fusarium of filamentous fungi is distributed worldwide and comprises at least 300 species [4,5]. First described by Link in 1809, Fusarium includes several important and well-known mycotoxigenic plant pathogens [4,6]. Additionally, this was the first fungal genus associated with ice nucleation activity (INA) [7]. INA is the ability to initiate water crystallization at temperatures above –38℃, the freezing point of water in its ultrapure state [8]. Ice-nucleating particles (INPs) can be derived from a variety of sources, such as desert dust, anthropogenic emissions, marine, and biological aerosols. Biological INPs also include pollen, algae, lichens, and microorganisms like viruses, bacteria, and fungi [9]. In the case of Fusarium, INA has been reported above –12℃ for several species, including F. acuminatum, F. tricinctum, and F. avenaceum [10]. F. diversisporum was first described in rotten potato tubers in the US [11] and has been found in wheat fields in Iran [12]. However, limited information is available regarding F. diversisporum and its characteristics. No study has been published examining F. diversisporum as a phytopathogenic fungus. Furthermore, this species has not been reported in Korea. This study aimed to morphologically and molecularly characterize an unreported Fusarium species in Korea, F. diversisporum, isolated from a declined apple tree and determine its INA.
MATERIALS AND METHODS
Sample collection and fungal isolation
The fungal isolate KNUF-21-F39 was obtained from the branches of a declined apple tree (Malus domestica) collected from Chungcheongbuk province (36°33'50.0"N, 127°46'35.0"E), Korea. The strain was isolated and cultured on potato dextrose agar (PDA; Difco, Detroit, MI, USA) at 25℃. The strain was deposited into the general collection of the Korean Collection for Type Cultures (KCTC) (KCTC 56905).
Morphological characterization
The morphological characteristics of the designated strain KNUF-21-F39 were studied by culturing the strain on PDA and oatmeal agar (OA). The cultural characteristics of the strain and the colony color, texture, growth, shape, and size were examined after ten days of incubation at 25℃. The microscopic characteristics were observed using a light microscope (BX-50; Olympus, Tokyo, Japan) after 60 days.
Genomic DNA extraction, PCR amplification, and sequencing
Total genomic DNA of the fungal strain was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s instructions. The strain was identified using ITS1/ITS4 primers to amplify the internal transcribed spacer region of nrDNA (ITS) [13]. For the remaining partial gene sequences, EF1/EF2 were used as primers for the translation elongation factor 1-α (tef1) [14], RPB1-F5/RPB1-R8 and RPB1-F7/RPB1-G2r for the RNA polymerase largest subunit (rpb1) [15], and CAL-228f/CAL-2Rd for calmodulin (cal) [16,17]. The amplified PCR products were purified and sequenced (Bioneer Co., Daejeon, Korea), and the NCBI Basic Local Alignment Search Tool (BLAST) was used to search for sequence similarity percentages.
Phylogenetic analysis
The obtained sequences of ITS, tef1, rpb1, and cal were aligned with highly similar sequences identified using the BLAST search tool, and edited using BioEdit v.7.0.5.3 [18]. A phylogenetic tree was constructed based on the neighbor-joining method [19], as described by the Kimura model [20] using MEGA version X [21] with bootstrap values based on 1,000 replications.
Ice nucleation activity evaluation
The strain KNUF-21-F39 was grown on PDA and potato dextrose broth at 25℃ for seven days. Solid media-grown mycelium was removed from the PDA plates, suspended in 500 µL of sterile double-distilled water, and vigorously homogenized. Liquid cultures were filtered using an AVANTEC® 0.45 µm disposable membrane filter. A droplet freezing assay was conducted at –8℃ based on previous studies [7,22] as described by Avalos-Ruiz et al. (2022) [23]. Frozen droplets were observed visually and the strain was considered IN-active if 4/5 or 5/5 droplets froze within one minute. Double-distilled water was used as a negative control, and Pseudomonas syringae pv. syringae strain KACC 21200 was used as the positive control. INA was confirmed using a tube-freezing assay; the strain was considered positive if freezing was observed after two minutes [23].
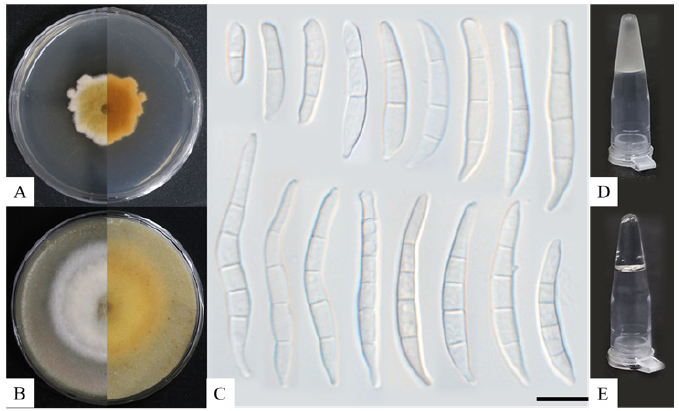
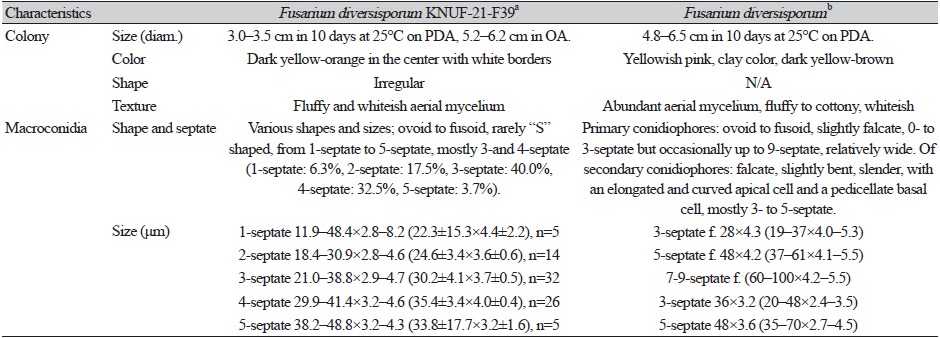
Fig. 1. Morphological characteristics and ice nucleation activity of Fusarium diversisporum KNUF-21-F39. A, Colony formation on potato dextrose agar (PDA) after ten days. B, Colony formation on oatmeal agar after ten days. C, Macroconidia observed after 60 days of growth in PDA. D, Ice nucleation activity of the strain observed using the tube-freezing essay. E, Ice nucleation negative control (without freezing). Scale bars: 10 μm.
RESULTS AND DISCUSSION
Fusarium diversisporum Sherb., Memoirs of the Cornell University Agricultural Experimental Station 6:161 (1915) [MB#194315] (Fig. 1)
Specimen examined: Boeun-gun, Chungcheongbuk-do (36°33'50.0"N, 127°46'35.0"E) isolated from a declined apple tree (Malus domestica) branch.
Morphological characteristics of KNUF-21-F39
The strain KNUF-21-F39 showed a slow growth rate, reaching colonies with diameters ranging from 30–35 mm after ten days of incubation on PDA at 25℃, and 52–62 mm after ten days on OA. The colonies on PDA were irregularly shaped and dark-yellow to orange in color, with white margins and whiteish aerial mycelium that gave the colonies a fluffy appearance (Fig. 1A). Colonies on OA had a white fluffy appearance, with a ring-like outer border (Fig. 1B). The morphology of the strain was compared to previous descriptions [24]. The results indicated that the strain KNUF-21-F39 had a slower growth rate; however, the colony characteristics on PDA were consistent with those observed in previous studies. Macroconidia presented several shapes and sizes, from ovoid to fusoid, 1- to 5- septate, with “S” -shaped macroconidia rarely observed. Macroconidia primarily presented 3- to 4-septate morphology (Fig. 1C). Sizes were 11.9–48.4×2.8–8.2 (22.3±15.3×4.4±2.2), n=5 (1-septate, 6.3%), 18.4–30.9×2.8–4.6 (24.6±3.4×3.6±0.6), n=14 (2-septate, 17.5%), 21.0–38.8×2.9–4.7 (30.2±4.1×3.7±0.5), n=32 (3-septate 40.0%), 29.9–41.4×3.2–4.6 (35.4±3.4×4.0±0.4), n=26 (4-septate, 32.5%), and 38.2–48.8×3.2–4.3 (33.8±17.7×3.2±1.6), n=5 (5-septate, 3.7%) (Table 1). No chlamydospores or microconidia were observed. The culture and morphological characteristics of the isolated strain KNUF-21-F39 suggested that the fungus was closely related to F. diversisporum.
Molecular phylogeny of KNUF-21-F39
Sequences containing 566, 594, 1510, and 588 bp were obtained that corresponded to the ITS region and partial sequences of tef1, rpb1, and cal genes, respectively. All obtained gene sequences were registered in the National Center for Biotechnology Information (NCBI) with the following GenBank accession numbers (ITS: OP113837, tef1: OP186041, rpb1: OP186042, cal: OP186043). BLAST results showed, for the ITS region, a 99.82% similarity with several strains of F. acuminatum, such as N-51-1 (MT566456), Scobey-MT (MK729582), and JAC13614 (MK432763); and F. tricinctum strains DSM100325_DISC_RLCS08 (MT453281) and QB23 (MT123097). A 99.46% of similarity was observed with F. diversisporum CBS 795.70 (=BBA 11129, MH859946). For the tef1 partial sequence, 100.0% similarity was observed with several F. avenaceum strains, such as Fv5-9 (MK111429), G17XY4-45 (MG670378) and JA-0925 (KP170732); 99.83% with F. tricinctum RTN17 (LC500690), and F. diversisporum BBA 11129 (MZ921930). For the rpb1 gene, 100.0% of similarity was observed with F. diversisporum BBA 11129 and 99.9% or less with several strains of F. avenaceum like LC6387 (MW024699) and LC6044 (MW024695).
Examination of the similarity of ITS, tef1, and rpb1 indicated that the isolate KNUF-21-F39 belonged to the genus Fusarium and, more specifically, to the F. tricinctum species complex. However, it was not possible to precisely identify the strain at the species level. For the cal gene, 100.0% similarity with F. diversisporum BBA 11129 (MZ921621) and 98.85% similarity with F. avenaceum ITEM7630 (LR215911) and ITEM13593 (LR215892) was observed, allowing the differentiation between F. avenaceum and F. diversisporum that was not attained before. The phylogenetic tree based on cal partial sequences showed that the strain KNUF-21-F39 clustered together with F. diversisporum BBA 11129 (Fig. 2). Therefore, the strain KNUF-21-F39 was identified as F. diversisporum.
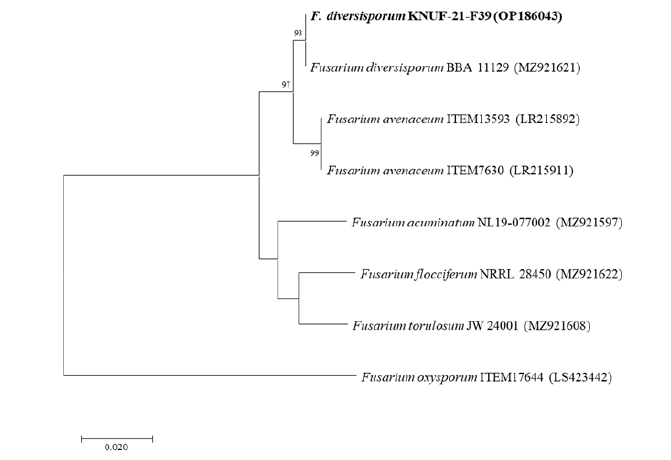
Fig. 2. Neighbor-joining phylogenetic tree based on cal partial sequences showing the position of Fusarium diversisporum KNUF-21-F39. F. oxysporum ITEM17644 was used as an out-group. The strain isolated in this study is marked in bold. Bootstrap values are based on 1,000 replications, and values below 70 are not shown. Bar, 0.02 substitutions per nucleotide position. Sequence accession numbers are presented in parentheses.
Ice nucleation activity of KNUF-21-F39
The strain showed high INA (5/5 frozen droplets) in both mycelial and mycelia-free suspensions (data not shown). INA was confirmed for KNUF-21-F39 using a tube-freezing assay, yielding a positive result for INA (Fig. 1D) compared to that observed with the negative control (Fig. 1E).
In 1992, two Fusarium species were reported to be IN-active: F. acuminatum and F. avenaceum [7]. Since then, F. oxysporum, F. tricinctum [25], F. armeniacum, F. begoniae, F. concentricum, and F. langsethiae [10] have been reported as IN-active species. However, information regarding the distribution and mechanisms of INA across Fusarium is lacking. In Korea, INA has been reported for microorganisms such as Pseudomonas syringae pv. syringae [26,27] and F. tricinctum [23]. In this study, the strain KNUF-21-F39 was collected from a declined apple tree (M. domestica) in Chungcheongbuk Province. The strain presented morphological and molecular characteristics similar to those previously reported for F. diversisporum. In addition, the strain presented the ability to act as an ice nucleator at −8℃.
Authors have suggested that the production of INA metabolites by some species of the genus Fusarium could be used for taxonomic classification [7]. Moreover, the production of INA metabolites can play an important environmental role and may be associated with the virulence and survival of the species [10]. Previous reports have stated that the INA of P. syringae can cause frost injury in sensitive plants [22]. Similarly, enhanced infection of plant roots by fusaria has been observed under freezing conditions [25], highlighting the importance of examining the INA of fungal species and strains along with their potential pathogenicity factors.
To our knowledge, this is the first report of F. diversisporum in Korea, and the first report of F. diversisporum as an IN-active fungus. With this study, we hope to expand the knowledge about fungal strains in Korea, as well as IN-fungal strains and their potential ecological roles.