INTRODUCTION
Tilia spp. (Malvaceae) are widely distributed throughout the temperate northern hemisphere including Asia, Europe, and eastern North America [1]. T. mandshurica Rupr. & Maxim., commonly named ‘mandshurian linden’, is a deciduous tree and native to northeastern Asia including China, Japan, Korea, and Russia (Siberia). In Korea, the tree is one of the most common trees in gardens and parks and a nectar source of small pollinators such as honeybees and flies [2].
On T. mandshurica, a rust fungus (Pucciniastrum tiliae) has been recorded from Japan and China, along with a dieback and canker-causing fungus (Nectria dematiosa from China) and two powdery mildew fungi (Erysiphe clintonii from USA and E. oleosa var. zhengii from Russia) [3]. Pucciniastrum tiliae is known to form spermogonial and aecial stages on Abies spp. but uredinial and telial stages on Tilia spp. [4].
After a rust disease was observed on T. mandshurica, planted in an experimental forest of Korea University (37°30'20"N; 127°41'56"E), Yangpyeong, Korea in August 2006, it has been consistently observed in Goseong, Hoengseong, and Incheon. The rust disease on T. mandshurica caused premature defoliation (Fig. 1A). The affected leaves were discoloured and became yellow. Uredinia were formed on the lower leaf surfaces (Figs. 1B and C). The present study aimed to record this new rust disease on T. mandshurica in Korea and to identify the causal fungus based on morphological characteristics and molecular sequence data.
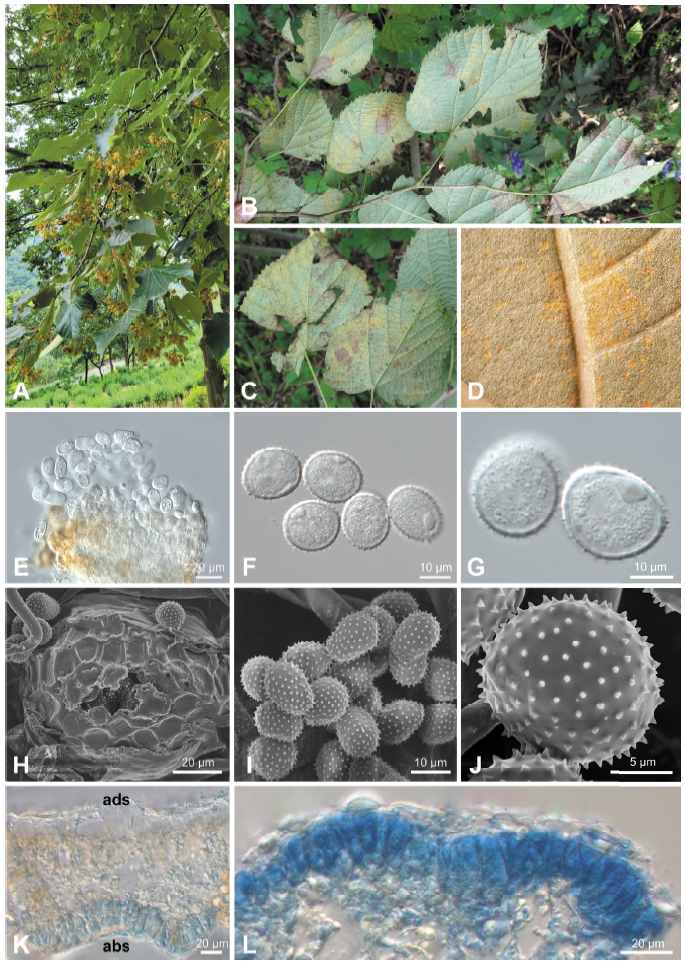
Fig. 1. Rust disease caused by Pucciniastrum tiliae on Tilia mandshurica . (A) Early defoliation of an affected tree. (B and C) Rust symptoms on the lower surface of leaves. (D) Uredinia formed on the lower surface of a leaf. (E) Uredinium under a differential interference contrast (DIC) microscope. (F and G) Urediniospores under a DIC microscope. (H) Uredinium under a scanning electron microscope (SEM). (I and J) Urediniospores under a SEM. (K) Vertical section of telium (abs, abaxial side; ads, adaxial side). (L) Teliospores in a leaf epidermis.
MATERIALS AND METHODS
For morphological investigation, rust-infected leaves were observed under a dissecting microscope (M205C; Leica, Wetzlar, Germany), a DIC light microscope (Axio Imager 2, Carl Zeiss, Oberkochen, Germany) and a scanning electron microscope (S-4800+EDS, Hitachi, Tokyo, Japan). All voucher specimens were deposited at the Korea University herbarium in Seoul, Korea. Information of all voucher specimens used for morphological and molecular phylogenetic analyses is provided in Table 1.
To confirm morphological identification, genomic DNA was extracted from urediniospores on infected leaves using MagListo 5M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The internal transcribed spacer (ITS) and large subunit (LSU) rDNA regions were amplified using primer pairs ITS5u/ITS4rust [5,6] and LRust1R/LRust3 [5], respectively. The PCR products were purified using AccuPrep® PCR/Gel Purification Kit (Bioneer, Daejeon, Korea) and sequenced by a DNA sequencing service (Macrogen, Seoul, Korea) with the primers used for amplification. The resulting sequences were edited using the DNASTAR software package (Lasergen, Madison, WI, USA) and deposited in GenBank (OL519187–OL519191 for ITS and OL519193–OL519197 for LSU). Phylogenetic trees were constructed by the maximum-likelihood (ML) method using MEGA 7 [7], with the default settings of the program, with the Tamura-Nei model. The robustness of individual branches was estimated by bootstrapping 1,000 replicates.
RESULTS AND DISCUSSION
Uredinia were hypophyllous, scattered to grouped, bright yellow or orange, round, covered by the epidermis (Fig. 1D), and 50-100 μm in diameter (Figs. 1E and H). Urediniospores were subglobose to ellipsoidal, pale yellow, and measured (17.0-)18.3-20.7(-21.5)×(14.4-)15.0-16.2(-17.7) μm (av. 19.6×15.5 μm), with a wall of echinulate ornamentation and 0.7-1.6 μm thick (Figs. 1F, G, I, and J). Telia were hypophyllous, subepidermal, scattered to grouped, and brown. Teliospores were formed under the host plant epidermis, divided into 2-4 cells, ellipsoidal, angular or oblong, and measured (23.8-)27.3-35.4(-38.3)×(4.7-)6.4-9.7(-11.5) μm (av. 31.4×8.0 μm) with a thin and smooth wall of 0.5-1.0 μm thick (Figs. 1K and L). These morphological features matched those reported for Pucciniastrum tiliae [4,8].
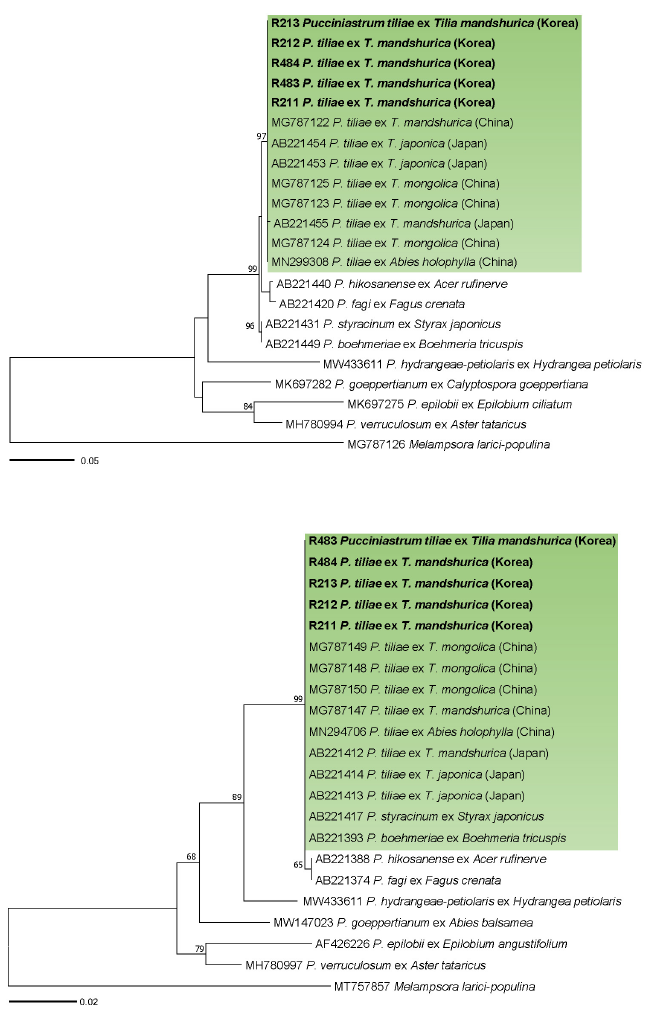
Fig. 2. Maximum likelihood tree of Pucciniastrum species inferred from the internal transcribed spacer (ITS) (A) and large subunit (LSU) (B) rDNA sequences. The numbers above the branches represent bootstrap values over 60%. The green boxes represent Pucciniastrum tiliae . The Korean specimens sequenced in the present study are shown in bold.
In BLASTn search, ITS rDNA sequences of the Korean specimens were identical with P. tiliae ex T. mongolica in China (MG787123, MG787124, MG787125), T. japonica in Japan (AB221453, AB221454), T. mandshurica in China (MG787122) and Abies holophylla in China (MN299308) but differed from P. tiliae ex T. mandshurica in Japan (AB221455) by only a nucleotide. LSU sequences revealed no nucleotide difference with all reference sequences of P. tiliae. In both maximum-likelihood phylogenetic trees of ITS (Fig. 2A) and LSU (Fig. 2B) sequences, the Korean specimens formed a well-supported clade with P. tiliae sequences ex T. mongolica, T. japonica, T. mandshurica, and Abies holophylla with high bootstrap values of 97% (ITS) and 99% (LSU).
Until now, P. tiliae had been recorded on eleven Tilia species, namely T. americana (Japan), T. amurensis (China, Japan, North Korea and Russia), T. japonica (Japan), T. kiusiana (Japan), T. laetevirens (China), T. mandshurica (China, Japan and Russia), T. maximowicziana (Japan), T. miqueliana (Japan), T. mongolica (China), T. platyphyllos (Japan) and T. tuana (China) [3,4,8]. Although Tilia spp. are widely distributed not only in Asia but also in Europe and Northeastern America, this rust fungus has yet been reported only in Northeast Asia. To our knowledge, this is the first report of P. tiliae on T. mandshurica in Korea. Since this pathogen has been previously recorded on T. amurensis in North Korea [8], and T. mandshurica in Northeast Asia (China, Japan, and Russia), the rust on T. mandshurica may have already existed in the Korean peninsula before the present study rather than newly emerging. The present study provides detailed information on morphological features and molecular data of this rust fungus on T. mandshurica. This finding will be useful for designing control measures against rust disease and conserving T. mandshurica.



