INTRODUCTION
Since the first report of indigenous yeast strains in 1910 [1], many studies on the excavation of yeast isolated from domestic materials have been actively conducted in Korea. From 1964 to 2020, 355 studies have reported on yeast and approximately 3,500 yeast strains have been preserved in the culture collections. These cultured yeasts comprise 500 species (including 9 variants) encompassing 142 genera and 48 classes of 2 phyla [2]. However, of these, 327 species from 114 genera are not cataloged in the National Species List of Korea (NSLK) as of December 2021 because there are no reports of taxonomic descriptions of these species, despite their extensive use in research and industry.
The NSLK is the backbone reference for claiming sovereign rights over biological resources, and its importance is growing with the adoption of the Nagoya Protocol. The yeast species Meyeromyma guilliermondii, Saccharomyces cerevisiae, Saccharomycopsis fibuligera, Wickerhamomyces anomalus, Candida tropicalis, and Papiliotrema flavescens are among the most 27 frequently reported species in literature, and numerous strains belonging to these six species are maintained in culture collections [2]. Therefore, the present study aimed to analyze the phylogenetic, morphological, and physiological characteristics of yeast strains belonging to these six species to list them in the NSLK.
MATERIALS AND METHODS
Species and strain selection
We listed 327 yeast species with no record in NSLK according to the number of publications or strains preserved at 9 Culture Collections in Korea [2]. Six extensively studied species with more than two strains stored in NIBR Culture Collections were selected. Thirty-one strains with diverse origin, such as isolation materials or geographic regions were finally chosen for taxonomic evaluation (Table 1).
DNA isolation, amplification and phylogenetic analysis
DNA was extracted from loopful yeast colonies obtained from fresh plate culture using a Nucleospin plant kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. The extracted DNA was used as a template in PCR amplification reactions with the primer pair NL1/NL4 [3] to amplify the D1/D2 domain of the large subunit of the rRNA gene (LSU). The amplicons were sequenced by Bioneer (Daejeon, Korea). Phylogenetic analysis was performed using D1/D2 region sequences and the phylogenetic tree was constructed with neighbor-joining methods based on the Tamura-Nei model using MEGA X software [4]. Bootstrap analysis was performed with 1,000 replicates. Taxonomic assignments were performed using the type strain of species within the genus or family.
Carbon source assimilation and oxidation
One representative strain of each species was selected for physiological experiments (carbon source assimilation and oxidation) and morphological observations. Carbon assimilation and oxidation tests were performed using a YT MicroPlate (Biolog, Hayward, CA, USA) consisting of 96 test wells, and each well was coated with 67 different carbon substrates. The assimilation of the carbon source was determined by an increase in turbidity. Oxidation was assessed by a color change from colorless to dark violet in positive cases. The results were analyzed using a microplate reader (Biolog, Hayward, CA, USA).
Morphology
Cell morphology was observed with cells grown on YM agar plate for 3 days at 25℃ with Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan). Colony form was observed with colonies grown for 7 days at 25℃. Hyphae and pseudohyphae formation were examined using Dalmau plates for 2 weeks on cornmeal agar at 25℃ following the standard protocol [5].
RESULTS AND DISCUSSION
In the present study, we performed taxonomic evaluations of 31 selected strains belonging to 6 species that frequently appeared in the literature or with available strains stored in the NIBR. These species included five ascomycetous yeasts (M. guilliermondii, S. cerevisiae, S. fibuligera, W. anomalus, and C. tropicalis) and one basidiomycetous yeast (P. flavescens). Strains with diverse habitats and geographic origins for each species were retrieved from the NIBR Culture Collection (Table 1). Phylogenetic analysis of the D1/D2 regions of the LSU rRNA gene from the selected strains was grouped with that of the (T) type strain of each species (Fig. 1A-6A). Five strains obtained from wild plants or soil formed a monophyletic group with M. guilliermondii type strain CBS2030 (Fig. 1A). The D1/D2 sequences of seven strains isolated from Nuruk, soil, and fruits or bark tissues of wild plants were identical to those of the neo-type (NT) strain of S. cerevisiae, NRRL Y-12632 (Fig. 2A), forming a single clade. Two strains from Nuruk were grouped with the type strain of S. fibuligera, Y-2388 (Fig. 3A), and eight strains from Nuruk were grouped with that of W. anomalus, CBS 5759 (Fig. 4A). Three strains isolated from soil samples had identical sequences in the D1/D2 region of LSU to those of the type strain of C. tropicalis NRRL Y-12968 (Fig. 5A). Six strains from wild plants and soil were grouped with the type strain of P. flavescens, CBS 942. The morphological and physiological characteristics of the selected strains of each species also supported the identification of each strain as a designated species. This study provides results supporting data for listing the six species in the NSLK.
Species description
Meyerozyma guilliermondii (Wick.) Kurtzman & M. Suzuki, Mycoscience 51: 7, 2010
The genus Meyerozyma (Debaryomycetaceae, Saccharomycetales) was first proposed based on phylogenetic analysis of the D1/D2 regions of LSU and SSU rRNA gene sequences to accommodate Pichia guilliermondii and P. caribbica [6]. Yurkov et al. [7] transferred five Candida species, C. athensensis, C. guilliermondii var. carpophila, C. elateridarum, C. neustonensis, and C. smithsonii to the genus Meyerozyma while adding M. amylolytica. Eight species are currently accepted in this genus. Meyerozyma is characterized by multilateral budding, the inability to ferment sugars, and CoQ-9 as a major ubiquinone [6]. M. guilliermondii is the type species of the genus Meyerozyma. In the present study, we report this species as an unrecorded species in South Korea.
Colonies are smooth and shiny ivory in color after 1 week on YM agar at 25℃. The cells are ovoid to elongate after 3 days on YM agar at 25℃, 2.0-4.1×3.2-5.1 μm, and occur singly or in pairs. Budding is by multilateral on a narrow base. After 2 weeks of culture on Dalmau plates at 25℃ (Fig. 1E), well-branched pseudohyphae but not true hyphae bearing whorls of blastoconidia are formed. Ascospores are not observed. This species is reported as heterothallic, following the pairing of complementary mating types, the resulting asci produce one to four hat-shaped ascospores [8].

Fig. 1.Phylogenetic tree and morphological characteristics of Meyerozyma guilliermondii. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of M. guilliermondii strains isolated from Korea. Bold means representative strain. B-E. Morphology of M. guilliermondii NIBRFGC000500301. B. Colony on YM agar 7 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Budding cells occurring in short chains on YM agar 7 days at 25℃. E. Pseudohyphae and blastoconidia formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 μm.
On the Biolog YT plate, the strain NIBRFGC000500301 is positive for the oxidation of D-Cellobiose, Maltose, D-melezitose, D-melibiose, D-raffinose, sucrose, D-trehalose, α-D-glucose, and D-galactose. Assimilation of carbon compounds: inulin, D-cellobiose, maltose, D-melezitose, D-melibiose (w), D-raffinose, sucrose, D-trehalose, N-acetyl-D-glucosamine, α-D-glucose, D-galactose, and D-mannitol (w). No growth occurs on L-malic acid, D-glucosamine, L-rhamnose, L-sorbose, α-methyl-D-glucoside, salicin, xylitol, i-erythritol, glycerol, L-arabinose, D-arabinose, D-ribose, or D-xylose (Table 2).

Fig. 2.Phylogenetic tree and morphological characteristics of Saccharomyces cerevisiae. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of S. cerevisiae strains isolated from Korea. Bold means representative strain. B-D. Morphology of S. cerevisiae NIBRFGC000502737. B. Colony on YM agar 7 days at 25℃. C. Unconjugated, persistent asci (arrowhead) after 7 days onY M agar at 25 ℃. D. Budding cells on YM broth 7 days at 25℃. Bars, 10 μm.
Examined strain: NIBRFGC000500301, Korea, Gurye-gun, 30 Jun. 2017, isolated from the fruit of Cornus officinalis.
Remarks: This species has been reported under the synonyms Candida guilliermondii and Pichia guilliermondii in Korea. Isolates were obtained from soil, plants, Meju, and humans. M. guilliermondii is distributed worldwide, including the USA, Brazil, Israel, Japan, and Korea.
Saccharomyces cerevisiae Meyen ex E.C. Hansen, Medd. Carlsberg Lab.: 29, 1883
The genus Saccharomyces (Saccharomycetaceae, Saccharomycetales) includes the most famous yeast, S. cerevisiae which is a key ingredient in baking, brewing, and winemaking. The Saccharomyces sensu stricto group comprises nine yeast species: Saccharomyces cerevisiae, S. paradoxus, S. uvarum, S. mikatae, S. kudriavzevii, S. arboricola, S. eubayanus, S. pastorianus, and S. jurei [9]. Although S. cerevisiae has been widely used industrially and has been researched since 1910 in Korea [1], this species has not been taxonomically described [2].
After 1 week on YM agar at 25℃, colonies are smooth, butyrous, and white to ivory colored. The cells are globose to broadly ellipsoid after 3 days on YM agar at 25℃, 3.3-5.6×4.7-8.3 μm, and usually occur singly or in pairs. Rudimentary pseudohyphae are occasionally formed after 2 weeks at 25℃, while septated hyphae are absent. Asci are persistent and containing one to four globose ascospores (Fig. 2C).
On the Biolog YT plate, the strain NIBRFGC000502737 is positive for the oxidation of D-melezitose, D-raffinose, Sucrose, α-D-glucose, and D-galactoseD-Galactose. But negative for D-cellobiose, Maltose, D-melibiose, or D-trehalose. Assimilation of carbon compounds: inulin, D-melezitose(w), D-raffinose, sucrose, α-D-glucose, D-galactose, and α-methyl-d-glucoside. No growth was observed on L-malic acid, D-cellobiose, maltose, D-melibiose, D-trehalose, N-acetyl-D-glucosamine, D-glucosamine, L-rhamnose, L-sorbose, salicin, D-mannitol, xylitol, i-erythritol, glycerol, L-arabinose, D-arabinose, D-ribose, or D-xylose (Table 2).
Examined strain: NIBRFGC000502737, Korea, Daegu, 10 Sep. 2018, isolated from the bark of Quercus acutissima.
Remarks: S. cerevisiae, the baker’s yeast, has been used for winemaking, baking, and brewing since ancient times and has been extensively studied in the food industry or as a model organism in biotechnology. This species is known as largely associated with fermented food and rarely found in the natural environment [10]. It has been reported as S. coreanus, the first reported yeast species in Korea [1], S. chevalieri, S. boulardii, S. capensis, S. diastaticus, S. italicus, S. cerevisiae var. ellipsoideus, S. ellipsoideus, S. fructuum, S. oviformis, S. steineri, and S. willianus. Although this species was more frequently isolated from fermented substances (Nuruk, Meju, and alcoholic beverages), many strains were also isolated from environmental samples such as soil, plants, seaweed, water, and even humans.
Saccharomycopsis fibuligera (Lindner) Klöcker, Die Gärungsorganismen in der Theorie und Praxis der Alkoholgarungsgewerbe: 299, 1924
Saccharomycopsis is the only genus in the family Saccharomycopsidaceae (Saccharomycetales), which was introduced by Schiӧnning [11]. The species delimitation of the genus Saccharomycopsis is unclear. Hajihosseinali et al. [12] reported 24 species in this genus whereas Yuan et al. [13] reported 19 species. Saccharomycopsis is characterized by multipolar budding, formation of true hyphae, and significant variations in the shape of ascospores (e.g., hat-shaped, reniform with terminal appendages, spherical or ellipsoidal, and have one or more ledges). S. phalluae has recently been added to the genus isolated from yellow rot lesions of Phallus rubrovolvatus in China [13].
After 1 week on YM agar at 25℃, aerobic growth is dull white and mycelial. The cells are ovoid to elongate after 3 days on YM agar at 25℃, 2.7-3.7×3.9-9.9 μm in size, sometimes tapered, and usually occur singly or in pairs. On Dalmau plate using cornmeal agar, pseudohyphae and true hyphae with varying numbers of blastoconidia are abundant. Asci are spherical to ovoid and each ascus forms two to four hat-shaped ascospores (Fig. 3E).
On the Biolog YT plate, strain NIBRFGC000134783 is positive for the oxidation of D-cellobiose, maltose, D-raffinose, sucrose, and α-D-glucose. But negative for D-melezitose, D-melibiose, D-trehalose, D-galactose. Assimilation of carbon compounds: inulin, D-cellobiose, maltose, D-raffinose, sucrose, α-D-glucose, i-erythritol, and glycerol. No growth on L-malic acid, D-melezitose, D-melibiose, D-trehalose, N-acetyl-D-glucosamine, D-glucosamine, D-galactose, L-rhamnose, L-sorbose, α-methyl-D-glucoside, salicin, D-mannitol, xylitol, L-arabinose, D-arabinose, D-ribose, or D-xylose (Table 2).
Examined strain: NIBRFGC000134783, Korea, Samcheok-si, 17 Jul. 2014, isolated from Nuruk.
Remarks: S. fibuligera is the major amyloytic yeast involved in food fermentation using rice and cassava [14]. It is usually isolated from highly starchy substrates worldwide whereas it is mainly isolated from Nuruk in Korea [2]. Endomycopsis fibuliger, isolated from red pepper paste [15], Saccharomyces fibuliger, and Pichia fibuligera are synonyms of this species. The ester-like odor and tufts of hyphal outgrowths on the colony surface are distinctive features of this species.
Wickerhamomyces anomalus (E.C. Hansen) Kurtzman, Robnett & Basehoar-Powers, FEMS Yeast Res. 8(6): 952, 2008
The genus Wickerhamomyces (Wickerhamomycetaceae, Saccharomycetales) was first proposed by Kurtzman et al. [16] in 2008 based on phylogenetic evidence. Nundaeng et al. [17] have re-evaluated the genus Wickerhamomyces accommodating 35 species with valid references. They proposed two novel species (W. lannaensis and W. nanensis) and a new combination (W. myanmarensis). In a recent study, W. sinyiensis was additionally proposed [18], resulting in 39 currently accepted species. Some species can utilize sugars for fermentation, and most species utilize diverse carbon sources, but not methanol or hexadecane. The predominant ubiquinone is CoQ-7 [16-17].
After 1 week on YM agar at 25℃, colonies are smooth, butyrous and white to tannish-white, margins entire. The cells are globose to elongate after 3 days on YM agar at 25℃, 2.5-5.0×3.5-7.7 μm, budding is multilateral on a narrow base, usually occur singly or in pairs (Fig. 4C). On Dalmau plate using cornmeal agar, pseudohypae are absent. Asci were not observed on YM agar and cornmeal agar after 1-2 weeks at 25℃.
On the Biolog YT plate, strain NIBRFGC000143644 is positive for oxidation of D-cellobiose (w), maltose, sucrose, α-D-glucose, and D-galactose. But negative for D-melezitose, D-melibiose, D-raffinose, or D-trehalose. Assimilation of carbon compounds: maltose, sucrose, and α-D-glucose. No growth on L-malic acid, Inulin, D-cellobiose, D-melezitose, D-melibiose, D-raffinose, D-trehalose, N-acetyl-D-glucosamine, D-glucosamine, D-galactose, L-rhamnose, L-sorbose, α-methyl-D-glucoside, salicin, D-mannitol, xylitol, i-erythritol, glycerol, L-arabinose, D-arabinose, D-ribose, or D-xylose (Table 2).
Examined strain: NIBRFGC000143644, Korea, Donghae-si, 12 May, 2015, isolated from Nuruk.
Remarks: W. anomalus is distributed worldwide and isolated from diverse substrates, such as soil, plants, food, and humans. Nuruk and other plant materials are major sources of this yeast species in Korea. There are many synonyms for W. anomalus, including Saccharomyces anomalus, the basionym, Endomyces anomalus, Pichia anomala, Willia anomala, and Hansenula anomala due to morphological and physiological variations among strains. Isolation of this species in Korea has been reported under the names H. anomala, H. anomala var. anomala, and P. anomala [2].

Fig. 3.Phylogenetic tree and morphological characteristics of Saccharomycopsis fibuligera. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of S. fibuligera strains isolated from Korea. Bold means representative strain. B-E. Morphology of S. fibuligera NIBRFGC000134783. B. Colony on YM agar 7 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Pseudohyphae bearing blastoconidia and asci on Dalmau plate with cornmeal agar for 2 weeks at 25℃. E. Hat shaped ascospores on cornmeal agar 2 weeks at 25℃. Bars, 10 μm.
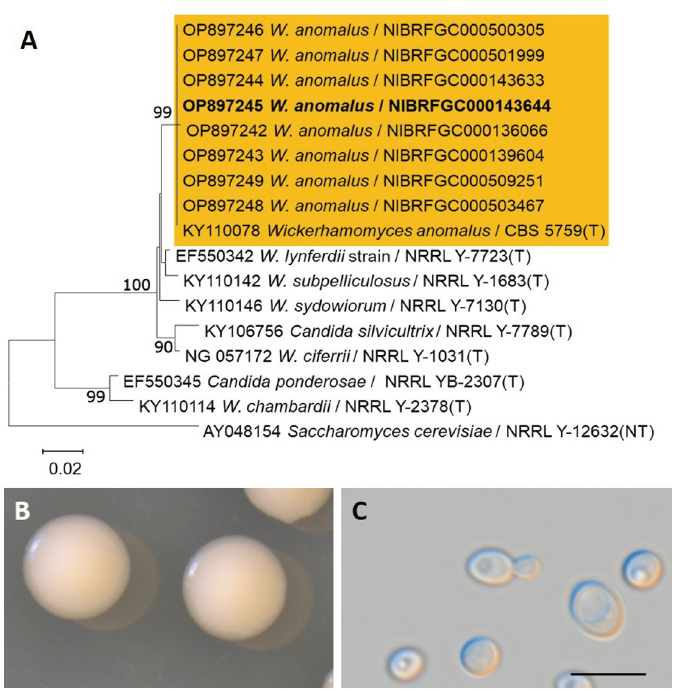
Fig. 4.Phylogenetic tree and morphological characteristics of Wickerhamomyces anomalus. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of W. anomalus strains isolated from Korea. Bold means representative strain. B-C. Morphology of W. anomalus NIBRFGC000143644. B. Colony on YM agar 7 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. Bars, 10 μm.
Candida tropicalis (Castell.) Berkhout, De schimmelgeslachten Monilia, Oidium, Oospora en Torula: 44, 1923
Candida (Saccharomycetales) is a highly polyphyletic, large, anamorphic genus [19]. Some Candida species can cause candidemia, the most common invasive bloodstream infection. Five species, Candida albicans, C. glabrata, C. tropicalis, C. parapsilosis, and C. krusei, account for 92% of candidemia cases [20]. Although C. tropicalis has been extensively studied for its clinical importance, it has also been studied for its phenol biodegradation [21] and ethanol or xylitol production [22]. Accurate identification of C. tropicalis is challenging because of its high degree of similarity to closely related species and the variability between strains. PCR-based methods have also been proposed for rapid and clear identification of this species [23].
After 1 week on YM agar at 25℃, colonies are smooth, butyrous and white in color. The cells are subglobose to ovoid after 3 days on YM agar at 25℃, 3.5-5.7×4.3-7.9 μm, and occur singly or in pairs. On Dalmau plates after 2 weeks at 25℃ (Fig. 5E), pseudohyphae with branched chains of cylindrical cells with blastoconidia are formed singly or in verticils, and true hyphae are present.

Fig. 5.Phylogenetic tree and morphological characteristics of Candida tropicalis. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of C. tropicalis strains isolated from Korea. Bold means representative strain. B-E. Morphology of C. tropicalis NIBRFGC000500169. B. Colony on YM agar 7 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Budding cells on YM broth 7 days. E. Pseudohyphae with blastoconidia on Dalmau plate with cornmeal agar. Bars, 10 μm.

Fig. 6.Phylogenetic tree and morphological characteristics of Papiliotrema flavescens. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2 domains of the large subunit (LSU) rRNA sequences, showing positions of P. flavescens strains isolated from Korea. Bold means representative strain. B-C. Morphology of P. flavescens NIBRFGC000502590. B. Colony on YM agar 7 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. Bar 10 μm.
On the Biolog YT plate, the strain NIBRFGC000500169 is positive for the oxidation of Maltose, D-melezitose (w), sucrose, D-trehalose, α-D-glucose, and D-galactose. But negative for D-cellobiose, D-melibiose, or D-raffinose. Assimilation of carbon compounds: inulin, maltose, D-melezitose(w), sucrose, D-trehalose, N-acetyl-D-glucosamine, α-D-glucose, and D-galactose. No growth on L-malic acid, D-cellobiose, D-melibiose, D-raffinose, D-glucosamine, L-rhamnose, L-sorbose, α-methyl-D-glucoside, Salicin, D-mannitol, xylitol, i-erythritol, glycerol, L-arabinose, D-arabinose, D-ribose, or D-xylose (Table 2).
Examined strain: NIBRFGC000500169, Korea, Daejeon, 9 Jan. 2017, isolated from soil around the Daejeon stream.
Remarks: C. tropicalis is a clinical yeast frequently encountered after infection with C. albicans, causing candidemia. This species appears to be distributed worldwide, including Jamaica, Brazil, Egypt, Italy, Russia, Japan, and Korea. It is isolated not only from clinical specimens but also from fruits and flowers of plants (Cactaceae), soil, water, and fermented drinks (kefir). In Korea, this species has been isolated from clinical, environmental (soil, water, and plant), and fermented samples (cheese, yogurt, Meju, and Nuruk).
Papiliotrema flavescens (Saito) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Studies in Mycology 81: 126, 2015
The genus Papiliotrema (Cryptococcaceae, Tremellales) was amended to accommodate 22 well-supported monophyletic species [24]. Subsequently, 6 new species were added to comprise 28 membered genus [25]. This genus is characterized by pale to brownish-colored colonies, absence of fermentation, and lack of nitrate utilization [26]. Although Papiliotrema flavescens has been recognized as a synonym of Cryptococcus laurentii (synonym of P. laurentii), differences in carbon utilization, whole-cell protein patterns [27], and rRNA gene sequences [28] indicate that these two species are separate species.
After 1 week on YM agar at 25℃, colonies are butyrous to mucoidal, smooth and entire margin, yellowish-cream in color. The cells are subglobose to fusoidal after 3 days on YM agar at 25℃, 3.5-5.5×4.4-6.8 μm, budding is lateral (Fig. 6C). On Dalmau plate using cornmeal agar pseudohyphae and true hyphae were absent. Asci were not observed on YM agar or cornmeal agar after 1-2 weeks at 25℃.
On the Biolog YT plate, the strain NIBRFGC000502590 is positive for the oxidation of D-cellobiose, maltose, D-melezitose, D-melibiose, D-raffinose, sucrose, D-trehalose, α-D-glucose, and D-galactose. Assimilation of carbon compounds: L-malic acid, inulin, D-cellobiose, D-melezitose, D-melibiose, D-raffinose, sucrose, D-trehalose, α-D-glucose, D-mannitol, and D-xylose. No growth on maltose, N-acetyl-D-glucosamine, D-glucosamine, D-galactose, L-rhamnose, L-sorbose, α-methyl-D-glucoside, salicin, xylitol, i-erythritol, glycerol, L-arabinose, D-arabinose, or D-ribose (Table 2).
Examined strain: NIBRFGC000502590, Korea, Geumsan-gun, 9 Jul. 2018, isolated from plant culture soil.
Remarks: This species was accommodated in the genus Papiliotrema after multigene phylogenetic analysis using the basionym Torula flavascens [24]. Cryptococcus flavescens, Rhodotorula flavescens, Torulopsis flavescens, and Cryptococcus laurentii var. flavescens are synonymous with those of P. flavescens. The major sources of Korean Papiliotrema strains are environmental substances, such as soil, plants, and water.