The family of Cornaceae comprises eight species and one variety from the genus Cornus L. in Korea: C. alba L., C. canadensis L., C. controversa Hemsl., C. florida L., C. kousa Bürger ex Hance, C. kousa subsp. chinensis (Osborn) Q.Y. Xiang, C. macrophylla Wall., C. officinalis Siebold & Zucc., and C. walteri Wangerin (List of vascular plants of the Korean Peninsula, 2021). Plants of this genus are commonly known as dogwood and are deciduous trees, which are mainly planted for horticultural and medicinal purposes. To date, two powdery mildew species have been associated with Cornus spp. in Korea: Phyllactinia corni H.D. Shin & M.J. Park on C. officinalis, and Microsphaera pulchra Cooke & Peck (now Erysiphe pulchra [Cooke & Peck] U. Braun & S. Takam.) on C. controversa and C. florida [1-3].
Previously, M. pulchra on C. macrophylla and C. controversa was treated as M. pulchra var. japonica (Henn.) U. Braun, based on slight morphological differences [4]. Later, this variety was reduced to the synonym E. pulchra [5]. Subsequently, two Erysiphe species were associated with Cornus spp. globally in the last decade: E. tortilis (Wallr.) Link. distributed in Europe and Iran onC. alba, C. amomum Mill., C. bretschneideri L. Henry, C. controversa, L., C. sanguinea L., C. sanguinea subsp. australis (C. A. Mey.) Soó, and C. sericea L.; and E. pulchra in Asia and North America on C. alternifolia L.f., C. controversa, C. florida L., C. kousa, and C. macrophylla [5]. Recent molecular-phylogenetic studies revealed that the powdery mildew on C. controversa is not E. pulchra, but E. cornicola Meeboon & S. Takam. The host range of E. pulchra is confined to C. florida and C. kousa [6].
During our extensive field forays, we repeatedly collected powdery mildew on C. controversa and preserved 29 samples in the herbarium of the Korea University (KUS), Seoul, Korea. The samples are labeled as KUS-F10074 (Oct 8, 1987, Chuncheon), F11814 (Aug 1, 1992, Gangneung), F12006 (Sep 20, 1992, Hongcheon), F12439 (Jul 2, 1993, Gangneung), F12477 (Jul 23, 1993, Gangneung), F13094 (Sep 28, 1994, Gangneung), F14045 (Aug 26, 1997, Gangneung), F15273 (Sep 25, 1998, Pyeongchang), F15473 (Oct 12, 1998, Seoul), F20629 (Aug 21, 2004, Hoengseong), F20928 (Nov 3, 2004, Pocheon), F23014 (Oct 15, 2007, Hoengseong), F24997 (Jun 25, 2010, Yangpyeong), F25605 (Nov 1, 2010, Yangpyeong), F25834 (Nov 21, 2011, Hongcheon), F26033 (Aug 29, 2011, Gangneung), F26856 (Aug 27, 2012, Inje), F27330 (Jul 3, 2013, Gapyeong), F27565 (Sep 5, 2013, Gapyeong), F28720 (Jul 6, 2015, Chuncheon), F29202 (Jun 8, 2016, Namyangju), F29897 (Aug 28, 2017, Namyangju), F31040 (Jun 24, 2019, Yangpyeong), F31405 (Nov 4, 2019, Gapyeong), F32924 (Jun 7, 2022, Wanju), F32956 (Jun 15, 2022, Wanju), F32972 (Jun 20, 2022, Buan), F33069 (Jul 17, 2022, Wanju), and F33301 (Sep 29, 2022, Imsil).
For morphological examination, asexual and sexual diagnostic structures were detached from powdery mildew colonies on fresh leaves, mounted in a drop of distilled water or 3% NaOH solution on a glass slide, and examined under an optical microscope using bright-field and differential interference contrast. At least 20 measurements of each structure were taken using an Olympus BX51 microscope (Olympus, Tokyo, Japan) at 100× and 1,000× magnifications, and a Zeiss AX10 microscope equipped with an AxioCam MRc5 (Carl Zeiss, Göttingen, Germany) was used to take photographs. The morphological description was based mainly on representative samples of KUS-F32956 accommodating the anamorph and F31405 holding the teleomorph.
Mycelia on both sides of the leaves, herbaceous stems, and inflorescences (esp. floral axis) were persistent to evanescent, forming circular to irregular white patches, or covering the entire surface with a thin layer of mycelium (Fig. 1A). In autumn, chasmothecia appeared on the leaves, abundantly on the lower leaf surface, and on the herbaceous stems (Figs. 1B and C). Hyphal appressoria were well-developed, moderately lobed to multilobed, and single or in pairs (Figs. 1D and E). Conidiophores were single, or occasionally two or three were observed on a hyphal cell, arising from the upper part of the mother cells, producing conidia singly, 56-90 µm long. Foot-cells were straight or slightly flexuous, 18-26×6-8 µm, simple, with a basal septum at the branching point or slightly displaced (Figs. 1F-L). Immature conidiophores with immature primary conidia were characterized by long foot cells (occupying approximately two-thirds of the full length) (Figs. 1F-H). Conidia were oval to ellipsoid, 22-30×13-17 µm (length/width ratio of 1.5-2.1), containing small oil drops, devoid of conspicuous fibrosin bodies, and producing germ tubes on the perihilar position (Figs. 1O-R). The primary conidia were generally smaller than the conidia, rounded at the apex, and subtruncated at the base (Figs. 1M and N).
Chasmothecia were scattered, rarely gregarious, 110-134 μm in diam., and globose to subglobose (Fig. 1S). Peridium cells were irregularly polygonal, conspicuous, and 12-24 μm in diam. Appendages were 10-18 in number, arising around the equatorial zone of the chasmothecia, stiff, straight to slightly bent, and dichotomously branched 3-5 times. Ultimate tips were recurved, 0.8-1.2 times as long as the chasmothecial diameter, 6-8 μm wide, hyaline throughout or pale brown at the base, thick-walled at the base and thinner upwards, smooth throughout, and aseptate or uniseptate at the base (Figs. 1T-V). Ascus were 3-7 in a chasmothecium, subglobose to broadly ellipsoid, 54-66×44-50 μm, short-stalked, wall 2-3 μm thick, terminal oculus thinner, and 8-spored (Fig. 1W). Ascospores were mostly oval to ellipsoid, 14-17×9-12 μm with a length/width ratio of 1.3-1.7, and colorless (Fig. 1W). These morphological features were compatible with those of Erysiphe cornicola Meeboon & S. Takam. [6].
Three specimens (KUS-F31040, F32956, and F32972) were used for molecular phylogenetic analysis. Total genomic DNA was extracted from mycelia using MaglistoTM 5M kits (Bioneer, Daejeon, Korea), according to the manufacturer’s guidelines. The primer pairs PM3/TW14 and ITS1/PM6 were used to determine the nucleotide sequences of the large subunit (LSU) gene and the internal transcribed spacer regions (ITS1 and ITS2) of the rDNA, respectively [7]. Six newly obtained sequences were assembled and deposited in GenBank under the accession numbers OQ061268, OQ061269, and OQ061271 for ITS and OQ061270, OQ067228, and OQ067229 for LSU. Subsequently, these sequences were combined and aligned with 17 closely related sequences of the genus Erysiphe using MUSCLE in MEGA 11 [8]. Erysiphe nothofagi (Thaxt.) U. Braun & S. Takam. (AB378746) was selected as the outgroup. Phylogenetic analysis was conducted based on a combined ITS and LSU dataset in PAUP* 4.0, using the maximum parsimony (MP) method [9]. The robustness of the internal branches was evaluated using bootstrap analysis with 1000 replicates. Tree scores, including tree length, consistency index, retention index, and rescaled consistency index, (RC) were calculated.
A BLAST search revealed that our sequences were 100% identical to those of E. cornicola (AB000941, AB015924, and LC228567) for ITS, whereas there was only one bp difference (910 bp out of 911; 99.89%) for the LSU gene with the sequence of E. cornicola (AB022389). In total, 20 sequences and 1,556 characters were used in the MP analysis, of which 1,371 (88.1%) characters were constant, 73 (4.7%) were parsimony informative, and 112 (7.2%) were variable or parsimony uninformative. In the resulting maximum parsimony tree, sequences of Korean isolates were clustered in a separate clade with sequences of E. cornicola and supported by a 100% bootstrap value (Fig. 2). This study provided detailed morphological characteristics of both the anamorph and teleomorph of E. cornicola based on Korean isolates and confirmed the identity of E. cornicola as being distinct from E. pulchra s. str. This is the first report of E. cornicola in Korea.

Fig. 1.Erysiphe cornicola powdery mildew on Cornus controversa . A: The powdery mildew infects the young leaves of Cornus controversa . B: Numerous chasmothecia formed on the lower leaf surface. C: Close-up view of chasmothecia formed on the leaf. D, E: Multilobed appressoria (arrows) formed on the hyphae. F-H. Young conidiophores. Note the round apex and long foot-cells. The arrow points to a slightly displaced basal septum. I-L: Conidiophores. Three conidiophores were formed on a hyphal cell (L). M, N: Primary conidia with apically rounded and basally subtruncate ends. O: Conidia. Note small oil drops. P-R: Conidia in germination. S: Chasmothecium with seven asci and 17 appendages. T-V: Chasmothecial appendages. W: Asci. Note eight ascospores each.
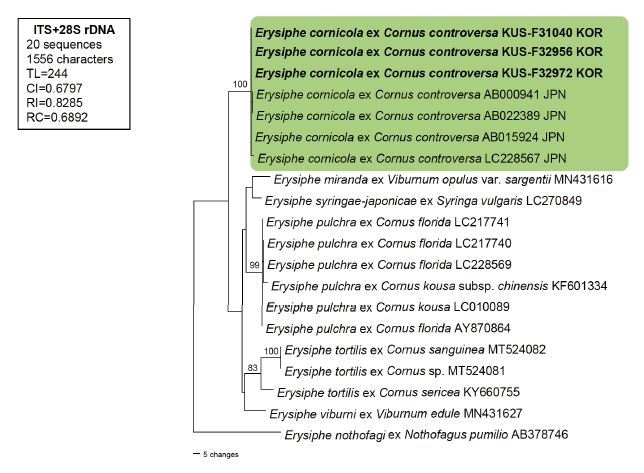
Fig. 2.A phylogenetic tree mainly representing Erysiphe spp. on Cornus spp., generated from a combined dataset of ITS and LSU genes of rDNA retrieved from 20 Erysiphe sequences based on the maximum parsimony method. The sequence of E. nothofagi (AB378746) represents an outgroup. The isolates obtained in this study are shown in bold. Bootstrap values (> 70%) are indicated on the related branches. Tree scores, such as tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) are presented in the box given on the left side of the Figure.


