Abstract
This study aimed to screen and investigate the microbiological characteristics, enzyme activity, and physiological functionality of unrecorded wild yeasts from the Wolsung valley of South Deogyu mountain and Jinjamcheon in Daejeon city, Korea.
Figures & Tables
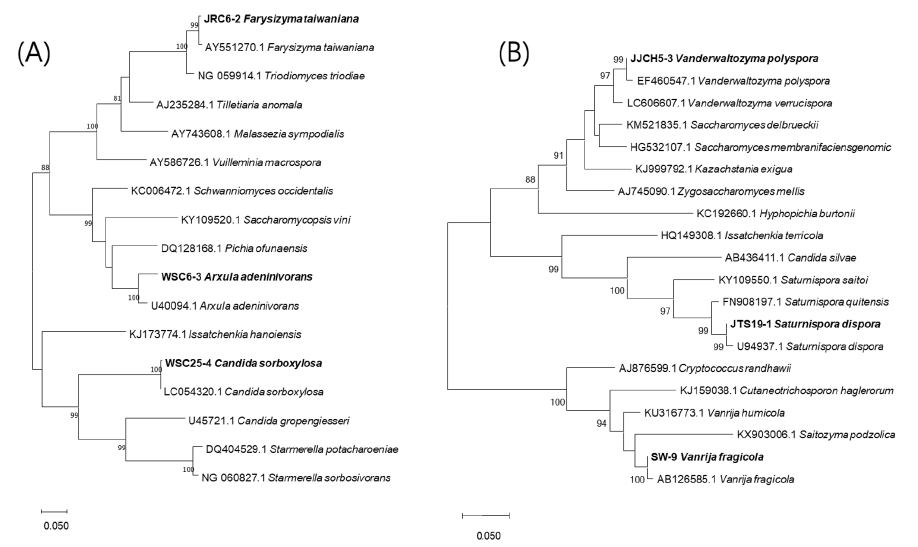
Fig. 1. Phylogenetic trees of unrecorded wild yeasts, JRC6-2, Arxula adeninivorans WSC6-3, Candida sorboxylosa WSC25-4 (A) and JJCH5-3, Saturnispora dispora JTS19-1, SW-9 (B), based on the sequences of D1/D2 region of the large-subunit rDNA. The tree was generated by the neighbor-joining method, using MEGA X. The bar indicates the number of substitutions per position. (Bold : unrecorded yeasts).


