INTRODUCTION
Ascomycetes belongs to the phylum Ascomycota, the largest fungal phylum, and consists of 93,000 species [1,2]. Ascomycota are widely distributed in various terrestrial, freshwater, and marine environments [3]. Although some ascomycetes live in soil or dung, most are saprobes [4]. Some exist as infections that affect humans, animals, and plants. Others live as parasites, such as endophytes or fungicolous or parasitic fungi [5-7]. The fungal genus Clonostachys (teleomorph Bionectria) belongs to the suborder Sordariomycetes of the Bionectriaceae family in Ascomycota. Globally, there are several species of Clonostachys. They live on recently deceased trees and decompose leaves as saprotrophs or as harmful mycoparasites and lichenicoles [8]. The use of Clonostachys as a multipurpose biocontrol agent has increased because of its capacity to inhibit sporulation of plant pathogenic fungi (mycoparasites), colonize senescent and dead tissues, stimulate plant growth, and facilitate plant resistance [9].
Chrysosporium is a genus of hyaline hyphomycete fungi belonging to the division Ascomycota, class Euascomycetes, order Onygenales, and family Onygenaceae [10]. A filamentous keratinophilic fungus, Chrysosporium is frequently identified in rotting wood, soil, animal waste, freshwater and marine sediments, feathers from birds and reptiles, the skin and hair of mammals. It feeds on feathers and hair fragments that remain in the soil [11].
The purpose of this study was to investigate recently discovered fungus species in Korea based on cultural and morphological characteristics, as well as their molecular phylogeny. The two fungal species are described and illustrated as a new record for the country of Korea.
MATERIAL AND METHODS
Sample collection and fungal isolation
The fungal isolates used in this investigation were present in soil samples collected from Cheongdo in Gyeongbuk Province (35°36′26.3″N, 128°40′21.5″E) and Dokdo Island (37°14′28.9″N, 131°51′54.5″E) in Korea. Soil samples were collected from the field at a depth of 15-30 cm using a pre-autoclaved sterile spatula, air-dried, and stored at 4℃ in a plastic bag. Fungal isolates were obtained using a traditional dilution plating method [12]. Single colonies were incubated for 4-5 days at 25℃ after transfer to potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates. The fungal isolates KNUF-20-NI011 and KNUF20-NI006 were chosen for additional molecular analyses and cultural and morphological characteristics. Fungal isolates were stored in 20% glycerol at -80℃ for further study. These strains have been deposited at the National Institute of Biological Resources (NIBR), with the accession number NIBRFGC000507832 and NIBRFGC000507846.
Cultural and morphological characterization
Cultural and morphological characteristics of the fungal isolates KNUF-20-NI011 and KNUF-20NI006 were recorded using different media, including PDA, 2% malt extract agar (MEA; Difco, Detroit, MI, USA), oatmeal agar (OA; Difco, Detroit, MI, USA), and phytone yeast extract agar (PYE; Merck KGaA, Darmstadt, Germany) with incubation for 7-21 days at 25℃ [13-15]. The growth of the fungi were quantified, and details of the colony, including its color, shape, and size, were noted. A light microscope (BX-50; Olympus, Tokyo, Japan) was used to investigate the morphological properties.
Genomic DNA extraction, PCR amplification, and sequencing
Total genomic DNA from the fungal isolates KNUF-20-NI011 and KNUF-20-NI006 were extracted from the fungal mycelia grown on the PDA plate using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s protocol. For the polymerase chain reaction (PCRmax, Alpha Cycler AC-1, Staffordshire, UK), the ITS1F and ITS4 primer pair was used to amplify the internal transcribed spacer (ITS) regions [16,17], primers NL1 and NL4 were used for partial gene sequences of the 28S rDNA large subunit (LSU) [18], and a fragment of β-tubulin (TUB2) was amplified using the primers T1 and T22 [19]. PCR amplification protocols were performed with slight modification as described [20]. The quality of PCR products was evaluated by electrophoresis on 1.2% agarose gels stained with ethidium bromide. Then PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by SolGent (Daejeon, Korea).
Molecular phylogenetic analyses
Phylogenetic analyses were performed using sequences retrieved from the National Center for Biotechnology Information (NCBI) (Table 1). The recovered sequences were aligned using the program Clustal X and Kimura’s two-parameter model, ambiguous regions were removed from the alignments, and evolutionary distance matrices were computed for the neighbor-joining (NJ) method [21]. The NJ method [22] was used to deduce tree topology using the MEGA7.0 software with bootstrap values based on 1,000 replications [23].
RESULTS
Morphology of isolate KNUF-20-NI011
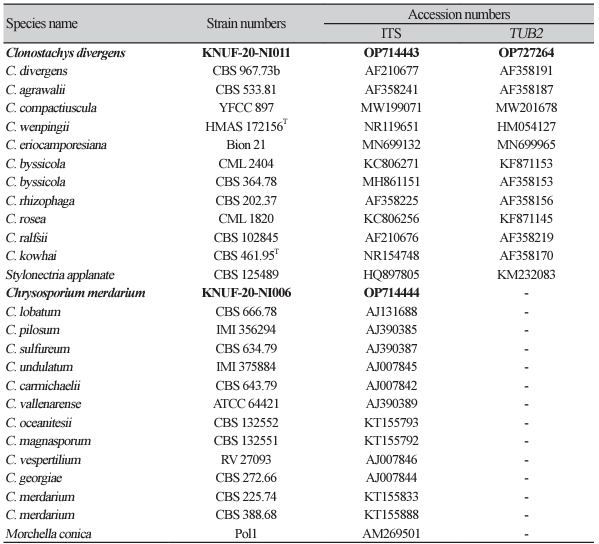
The colonies achieved a diameter of 32-33 mm in 14 days at 25℃ on MEA, whereas at 35℃, cultures did not exhibit any growth. An average growth of 34-34 mm in diameter was observed on PDA after 14 days at 25℃. In PDA cultures incubated in the dark, the reverse appeared strong yellow to brown within the center, pale orange, and the obverse was white, cottony to felty because of aerial mycelium; similar characteristics were observed in colonies cultured on MEA (Figs. 1A and C). Colonies on OA, the mycelium changed color to pale orange during incubation at room temperature after incubation at 25℃ in the dark. On OA medium, the reverse appeared light yellow, and the obverse was white and felty owing to strands of aerial mycelium or granular because of sporulation (Fig. 1B). Conidiophores were branches, strongly divergent, and almost rectangular. Phialides were divergent, young sporodochial pustules, young pustules had separate conidial chains formed by each phialide, collapsing to slimy masses (Figs. 1D and E). The conidia were ellipsoidal, with a flat hilum, slightly curved with one side slightly flattened, and had a diameter of 6.3-6.8×3.4-3.8 μm (Fig. 1F). The morphological and cultural characteristics of isolate KNUF20-NI011 revealed that it was mostly similar to previously identified Clonostachys divergens (Table 2) [8].
Fig. 1
Cultural and morphological characteristics of KNUF-20-NI011. Colony growing on potato dextrose (A), oatmeal (B), and malt extract agar (C) for 14 days at 25℃; Conidiophores with adpressed branches and phialides (D, E); Conidia (F). Scale bars: D-F=10 µm. Arrows indicate conidiophores and phialides.
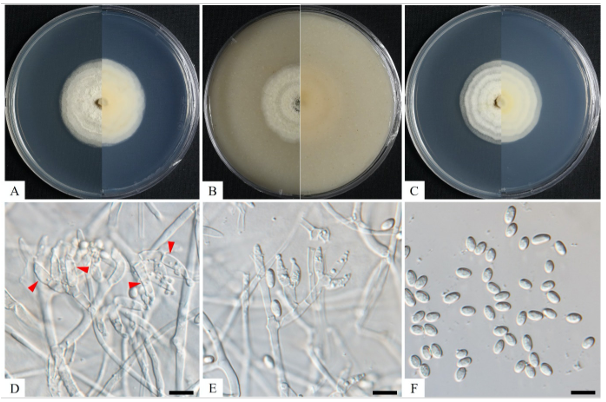
Molecular phylogeny of isolate KNUF-20-NI011
Through sequence analysis, 582 and 767 bp were obtained from the ITS regions and TUB2 portion, respectively. The BLAST results of ITS regions and TUB2 sequences exhibited a similarity of 99.3% and 98.5%, respectively, with different strains of Clonostachys divergens (CBS 967.73b). Based on NJ of the phylogenetic tree (combination of ITS regions and TUB2 sequences), isolate KNUF-20-NI011 clustered together with C. divergens (CBS 967.73b) with a bootstrap value of 100% (Fig. 2). Thus, isolate KNUF-20NI011 was identified as C. divergens, which is supported by the topology of the NJ phylogenetic tree.
Fig. 2
Neighbor-joining phylogenetic tree of KNUF-20-NI011 based on internal transcribed spacer (ITS) and β-tubulin (TUB2) sequences, demonstrating the phylogenetic position among the related strains in Clonostachys. Stylonectria norvegica CBS 139239 was used as an outgroup. The numbers above the branches represent the bootstrap values (>50%) obtained for 1,000 replicates. The strain isolated in this study is indicated in bold. Bar, 0.02 substitutions per nucleotide position.
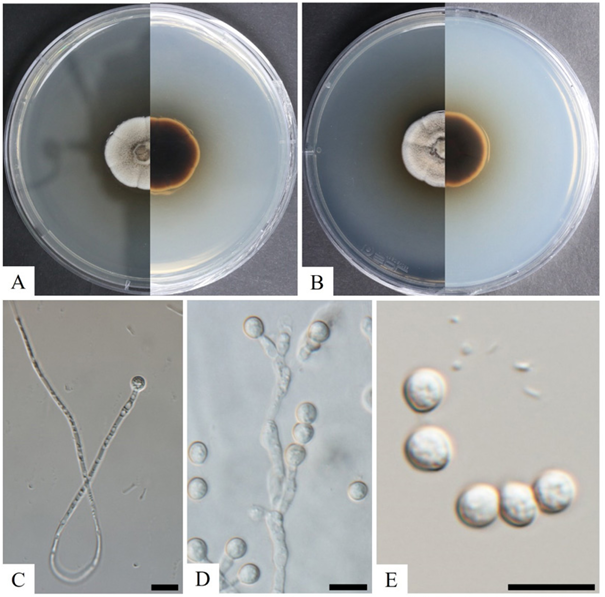
Morphology of isolate KNUF-20-NI006
The colony exhibited growth rates with the diameters of 30 mm and 31 mm after 14 days of incubation at 25℃ on PDA and MEA, respectively (Fig. 3). The colonies on PDA were yellowish brown in the center with undulated creamy-white margin and umbonate elevation surface; the reverse was black in the middle to creamy-brown (Fig. 3A). On MEA, the colony color ranged from cream to woody-brown in the center, with a white undulate margin and umbonate elevation surface; the reverse was black in the center to brown in the margin (Fig. 3B). The colonies were granular, dense, white floccose zone; reverse yellowish from center after 14 days at 25℃ on PYE. Aleuriospores were formed on the side of the hyphae, demonstrating a long tail and branch type (Figs. 3C and D). Spores were hyaline, smooth to roughened in texture, subglobose to pyriform, with thick walls and a size of 3.0-5.0×3.0-6.0 µm (Fig. 3E). The morphological and cultural characteristics of isolate KNUF-20-NI006 revealed that it is closely related to Chrysosporium merdarium (Table 3) [15].
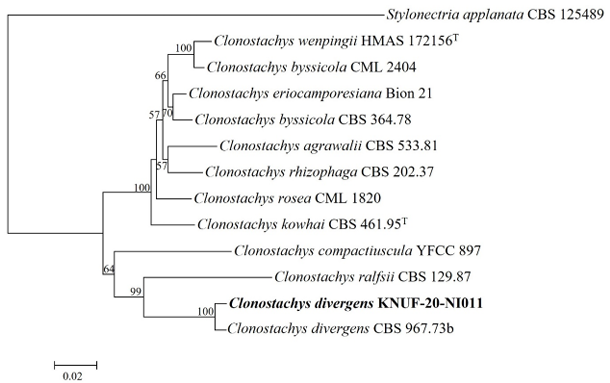
Molecular phylogeny of isolate KNUF-20-NI006
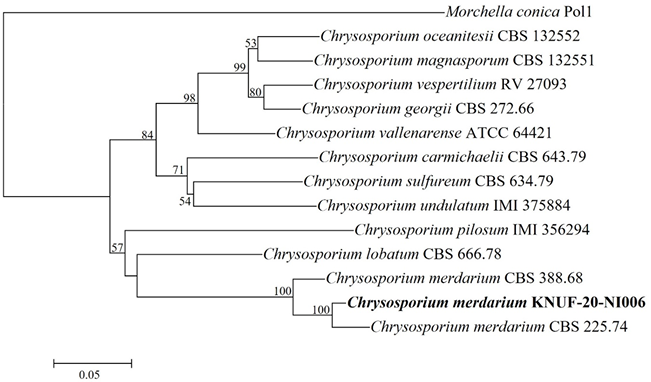
After sequencing analysis, 572 bp sequences were obtained from the ITS regions and the isolate KNUF-20NI006 revealed a high similarity of 99.6% with C. merdarium CBS 225.74 and 98.1% with C. merdarium CBS 388.68 based on 28S rDNA, whereas the ITS regions sequences shared 96.1% identity with C. merdarium CBS 225.74. The tree topology of isolate KNUF-20-NI006 is based on the concatenated ITS regions clustered among the strains of C. merdarium (CBS 338.68 and CBS 225.74) (Fig. 4). Thus, based on phylogenetic analyses, isolate KNUF-20-NI006 was identified as C. merdarium, a newly described fungus in Korea.
Fig. 4
Neighbor-joining phylogenetic tree of KNUF-20-NI006 based on internal transcribed spacer (ITS) sequences, demonstrating the phylogenetic position among the related strains in the genus Chrysosporium. Morchella conica Pol1 was used as an outgroup. The numbers above the branches represent the bootstrap values (>50%) obtained for 1,000 replicates. The strain isolated in this study is indicated in bold. Bar, 0.05 substitutions per nucleotide position.
DISCUSSION
Previous studies have reported that Clonostachys rosea was associated with avocado fruit rot in Puebla, Mexico [24]. Clonostachys ambigua and C. pallens were identified on bark (unknown host) from Indonesia [25]. Clonostachys species, such as C. rosea, are well-known biological control agents for various plant pathogens [26]. In the present study, the reported species, C. divergens (KNUF-20-NI011), was isolated from soil in Gyeongbuk Province, Korea.
Members of Chrysosporium are distributed worldwide and can produce many valuable metabolites, especially keratinase, which can be used widely in the chemical industry and in environmental protection, medicine, and agriculture [27,28]. Previously, C. vallenarense was obtained from the dung of the Arctic fox (Alopex lagopus) in Chile [29]. Chrysosporium sp. was reported to cause the death of rattlesnakes (Sistrurus catenatus) with severe facial swelling and disfiguration in Illinois, USA [30]. In 2006, C. linfenense was explored as a new species in the rhizosphere soil of Cedrus deodara in China [31]. Furthermore, the emergence of the keratinophilic fungus Chrysosporium (anamorph: Nannizziopsis vriesii) has caused fatal diseases in captive bearded dragons within the past decade [32]. In recent years, microorganisms have received increased attention owing to their negative impacts and crucial roles in agriculture, the chemical industry, and environmental protection. Additional research is required to determine the industrial significance and possible pathogenicity of the host species present under the ecological and environmental conditions of Korea. To the best of our knowledge, this is the first report of Clonostachys divergens and Chrysosporium merdarium in Korea.