INTRODUCTION
Approximately 2,700 fungal species from 240 genera are currently recognized in eight families in the order Hypocreales [1, 2]. The species produce light- to bright-colored, ostiolate, perithecial ascomata containing unitunicate asci with hyaline ascospores. The asexual morphs found in nature are, most frequently, moniliaceous and phialidic [3-7]. The fungal species are globally found in various environments; they are of great importance in agriculture and medicine and extensively exploited for industrial and commercial applications [5]. Moreover, several species have been reported as important opportunistic human pathogens [8-10], whereas others produce mycotoxins that are of medical concern [5].
The purpose of this study was to screen for unreported fungal species in field soil in Korea. This study provides information for further studies on the use of such organisms in agriculture and medicine as well as for industrial applications. On the basis of morphological and molecular characteristics, the strain KNU16-010, identified as Penicillifer diparietisporus, was reported for the first time in Korea.
MATERIALS AND METHODS
Collection of soil samples
In 2016, samples were collected from field soil during the screening of fungal species in Daegu, Korea (35°54′11.3″N, 128°36′56.1″E). A sterile trowel and spatula were used to collect the soil samples from a depth of 15~30 cm. The collected soil samples were placed in polythene zipper bags and transferred to the laboratory. Then, the soil samples were prepared for serial dilutions, and 1 g of soil was diluted with 10 mL of sterile distilled water. The serial dilutions were performed until a concentration of 10-3 was achieved, and, then, 100 µL of each sample was spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated for 2~3 days at 25°C. Single germinating fungal colonies were transferred to fresh PDA plates and incubated under the same conditions. The strain KNU16-10 was selected for further morphological and molecular phylogenetic analyses.
Morphological characterization
To study the cultural and morphological characteristics, PDA, oatmeal agar (OA; Difco), and malt extract agar (MEA; Difco) were used; the incubation period was 10 days at 25°C. Then, cultural characteristics such as colony color, fungal growth, and texture were observed and recorded, and the morphological characteristics were observed under a light microscope (BX-50; Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from 7-day-old colonies of the strain KNU16-010 grown on PDA by using the HiGene Genomic DNA Prep kit (BIOFACT, Daejeon, Korea), according to the manufacturer’s instructions. Partial gene sequences were amplified by the internal transcribed spacer (ITS) region, ITS1/ITS4, and translation elongation factor (TEF) 1-α gene, EF1-728F/EF2, as described previously [11-13]. Then, the amplified PCR products were purified with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Solgent (Daejeon, Korea). The similarities of the sequences were analyzed using BLAST of NCBI. The sequences obtained from KNU16-010 were deposited in NCBI GenBank (accession numbers LC387549 and LC387601 for the ITS region and TEF gene, respectively).
Phylogenetic analysis
The consensus sequences were compared with other sequences in the NCBI database by using BLAST to determine the percentage of shared sequence identity with other sequences of fungal species. The alignments were performed using MEGA 7.0 [14] with 1,000 bootstrap replicates, and the evolutionary distance matrices were generated based on Kimura’s neighbor-joining algorithm model [15].
RESULTS AND DISCUSSION
Morphology of the strain KNU16-010
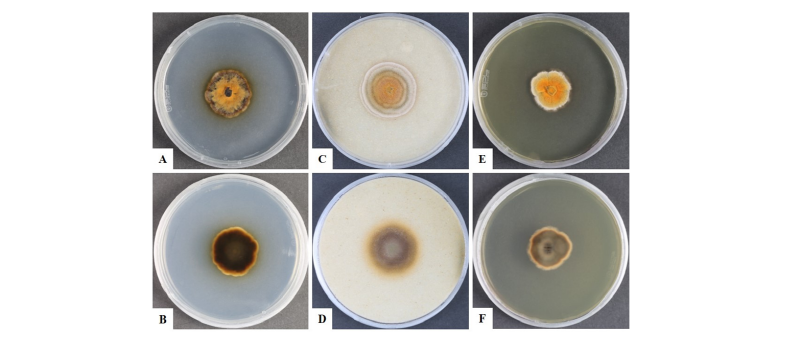
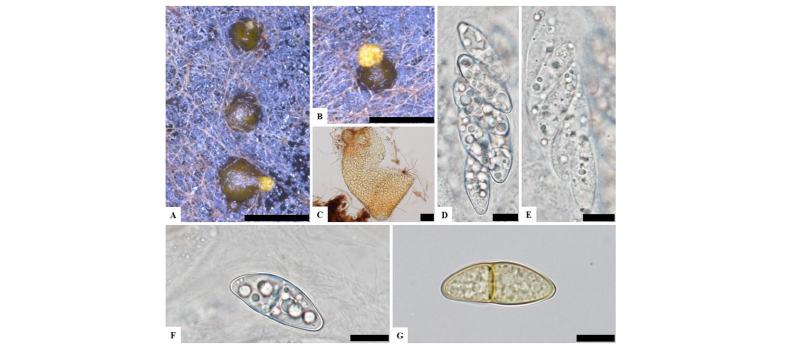
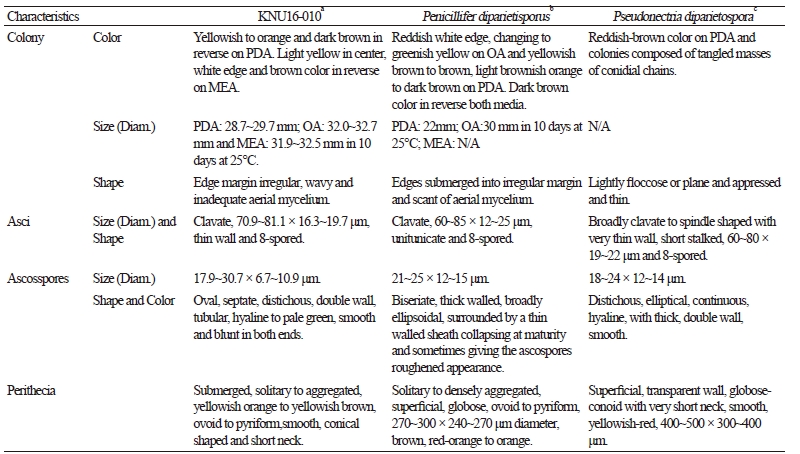
To observe the morphological characteristics, the strain KNU16-010 was cultured on PDA, OA, and MEA at 25°C for 10 days, and the diameter of the colonies was 28.7~29.7 mm, 32.0~32.7 mm, and 31.9~32.5 mm, respectively (Fig. 1). The morphology of the strain KNU16-010 was compared with previous descriptions of Penicillifer diparietisporus [16] and Pseudonectria diparietospora [17] (Table 1). Pseudonectria diparietospora produced reddish brown colonies on PDA [17]. Moreover, Neocosmospora arxii (basionym: Pseudonectria diparietispora) colonies are composed of hyaline or sometimes yellow-pigmented, branched, septate, smooth-walled, hyphae that are 2.5~5 μm wide, and they produce semi-immersed to immersed perithecia, scattered or aggregated in small groups, yellowish orange to yellowish brown, broadly ovoid to somewhat pyriform, 220~340 × 160~270 μm in diameter, almost glabrous, conical shaped with short neck [18]. However, our strain formed orange brown to yellowish red colonies in all the media, and PDA was used to observe the mycological characteristics (Fig. 1). The fungal colonies showed wavy mycelia at the edges and rough wart-like structures. They also formed septate, irregular, and short hyphae, and the average width of the hyphae was 2.42 μm. They produced submerged perithecia, solitary to aggregated, yellowish orange to yellowish brown, ovoid to pyriform, smooth, conical shaped with short neck (Fig. 2A, 2C). Asci clavate, 70.9~81.1 × 16.3~19.7 μm, thin wall, and 8-spored (Fig. 2D, 2E). Ascospores were oval, septate, guttulate, 2-celled, distichous, double wall, biseriate, tubular, hyaline and pale green, smooth and blunt at both ends with a diameter (n = 30) of 17.9~30.7 × 6.7~10.9 μm and no conidia (Fig. 2F, 2G). The ascospores were distichous, elliptical, continuous, hyaline, with thick, double wall, smooth 18~24 × 12~14 μm and no conidia [17]. Thus, these morphological characteristics of the strain KNU16-010 were very similar to those of Penicillifer diparietisporus.
Notes: Currently, the genus Viridispora is composed of four species, Viridispora diparietispora (= Penicillifer furcatus), Viridispora alata (= Penicillifer bipapillatus), Viridispora fragariae (= Penicillifer fragariae), and Viridispora penicilliferi (= Penicillifer macrosporus), each with its own Penicillifer asexual morphs [7, 16, 19, 20]. Lombard et al. [21] reported that the epithet Pseudonectria diparietispora (1957) pre-dates that of Penicillifer furcatus (1991), and the basionyms are provided below.
Penicillifer diparietisporus (J.H. Miller, Giddens & A.A. Foster) Rossman, L. Lombard & Crous, comb. nov.
Basionym: Pseudonectria diparietispora J.H. Miller, Giddens & A.A. Foster, Mycologia 49:793. 1957.
≡ Neocosmospora diparietispora (J.H. Miller, Giddens & A.A. Foster) Rossman,
Samuels & Lowen, Mycologia 85:699. 1993.
≡ Viridispora diparietispora (J.H. Miller, Giddens & A.A. Foster) Samuels &
Rossman, Stud. Mycol. 42:167. 1999.
= Neocosmospora arxii Udagawa, Horie & P. Cannon, Sydowia 41:353. 1989.
= Neocosmospora endophytica Polishook, Bills & Rossman, Mycologia 83:798. 1991.
= Penicillifer furcatus Polishook, Bills & Rossman, Mycologia 83:798. 1991.
Molecular phylogeny of the strain KNU16-010
Genetic sequences of the ITS regions and TEF1-α were analyzed to determine the evolutionary relationships (Fig. 3) with the strains obtained from GenBank (Table 2). After analyzing the nucleotide sequences, 523 bp and 438 bp sequences were obtained from the ITS regions and TEF1-α, respectively. The BLAST results showed 100% similarity with Penicillifer diparietisporus (CBS 376.59) and 99% with Penicillifer diparietisporus (KM231861) in the ITS regions and TEF1-α, respectively. The combined BLAST results suggested that the strain KNU16-010 showed 100% similarity with the strain Viridispora diparietispora (= Penicillifer diparietisporus) (CBS 102797) and Penicillifer diparietisporus (CBS 376.59, NR154310). The phylogenetic tree was constructed on the basis of the ITS regions and TEF1-α to the strain Penicillifer diparietisporus [21]. The strain KNU16-010 was closely clustered with Penicillifer diparietisporus (CBS 376.59), with a 100% bootstrap value (Fig. 3). The phylogenetic results support the fact that the strain KNU16-010 is Penicillifer diparietisporus.
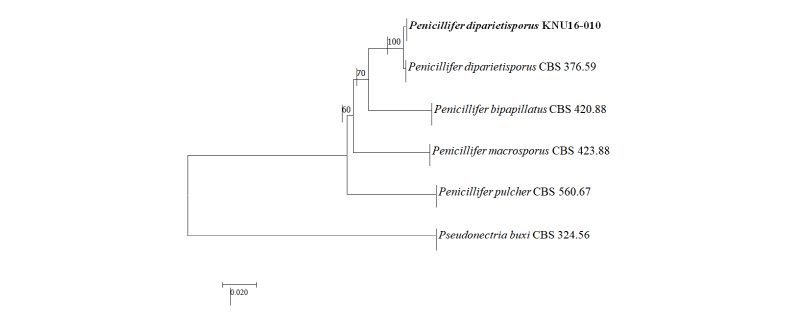
Fig. 3. Neighbor-joining phylogenetic tree based on the combined internal transcribed spacer sequences and translation elongation factor 1-α. Pseudonectria buxi CBS 324.56 was used as an outgroup. The strain isolated in this study is indicated in bold, and the bootstrap values are based on 1,000 replications. Bar, 0.02 substitutions per nucleotide position.
|
Table 2. GenBank accession numbers used in this study for the phylogenetic analyses 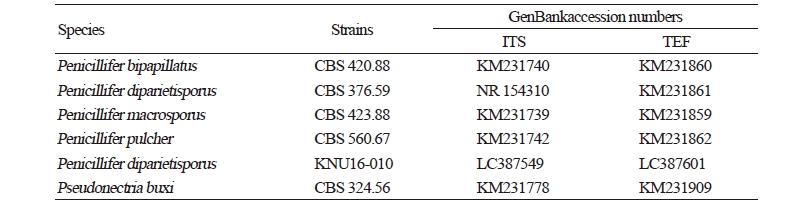
|
|
|
ITS, internal transcribed spacer; TEF, translation elongation factor. |
|
Members of the family Nectriaceae are pleomorphic fungi that display both asexual and sexual morphs during their life cycles [22]. In this study, we used molecular analysis, which provides a considerable challenge to conventional fungal systematics. Thus, more genomic studies of members of the family Nectriaceae are urgently required. To our knowledge, this is the first report of Penicillifer diparietisporus isolated from field soil in Korea.