INTRODUCTION
Endophytic fungi are asymptomatic organisms that live in healthy plant tissues throughout the entire lifecycle or part of the lifecycle [1]. Most are ascomycetes, and some are basidiomycetes [2, 3]. Many recent studies have focused on the biodiversity, secondary metabolites, and resistance to environmental stresses of these fungi [4, 5]. Moreover, various studies of endophytic fungi have demonstrated the potential of available secondary metabolites as bioactive substances [6].
Gymnosperms are distributed worldwide, with approximately 30~60 taxa, including cultivated or introduced species in Korea [7]. Among these taxa, only 3~6 conifer trees are endemic species in Korea [8, 9]. Current worldwide climate change and deforestation have threatened the survival of conifers, including those on the Korean peninsula. However, few studies have evaluated endophytic fungi that have evolved along with these trees in Korea. Therefore, it is necessary to investigate the biodiversity and ecological function of endophytic fungi in Korea.
In this study, we investigated the biodiversity of endophytic fungi in Juniperus rigida and Pinus densiflora from Mt. Baekryeonsan and Mt. Johangsan, Korea and compared the distributions of endophytic fungal communities according to host plant and locality.
MATERIALS AND METHODS
We collected twigs with healthy leaves from six individuals of J. rigida and 11 individuals of P. densiflora from Mt. Baekryeonsan (N38°27′, E126°39′) and three individuals of J. rigida and six individuals of P. densiflora from Mt. Johangsan (N38°27′, E126°39′). Collected leaves were subjected to surface sterilization. Surfaces of needle leaves were washed with tap water, and the leaves were treated using 1% NaOCl solution for 3 min and 70% ethanol for 2 min. The leaves were then washed with sterilized water twice, subjected to surface sterilization, and cut into 1 cm pieces. Four pieces were placed on potato dextrose agar (PDA) medium and incubated at 25°C in the dark. New hyphae were finally isolated [10].
Genomic DNA was extracted from the mycelia of isolates of endophytic fungi. DNA extraction was performed using a DNeasy Plant SV mini kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. Extracted DNA was amplified with primers ITS1F and ITS4 for the ITS region [11] and primers Bt2a and Bt2b for the β-tubulin region [12] using 2× Taq PCR Smart Mix2 (SolGent, Daejeon, Korea).
The polymerase chain reaction products were sequenced using a 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA) at SolGent, Korea. Analyzed sequences were identified on NCBI using BLAST, and neighbor-joining trees were generated using MEGA7 [13] to confirm the topological position among closest taxa with bootstrap analysis (1,000 replicates). Biodiversity and community structures of endophytic fungi were analyzed according to host plants and locality.
RESULTS AND DISCUSSION
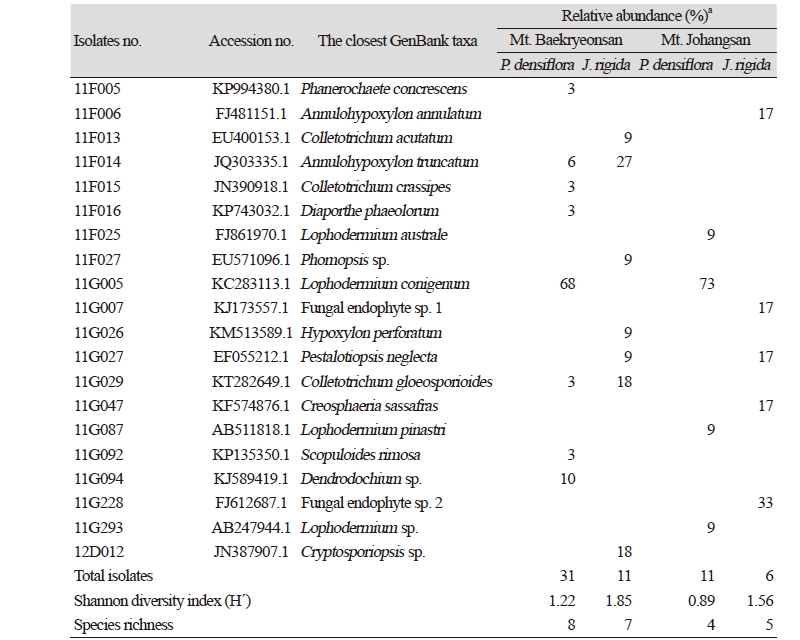
From 26 individual host plants, 59 morphotypes of endophytic fungi were isolated. The fungi were grouped into 20 taxa using sequence analysis (Table 1, Fig. 1). Most of the isolates showed similarities with the closest taxa above 96%. However, isolates 11F027, 11G007, 11G094, 11G228, 11G293, and 12D012 were not able to be identified at the species level. Fungal strains isolated from two species, P. densiflora and J. rigida, were divided into three classes: Leotiomycetes (62%), Sordariomycetes (34%), and Agaricomycetes (4%), similar to previous studies [14, 15]. Additionally, three species of endophytic fungi were identified for the first time in Korea. Their culture characteristics are described in Fig. 2, along with the phylogenetic analysis.
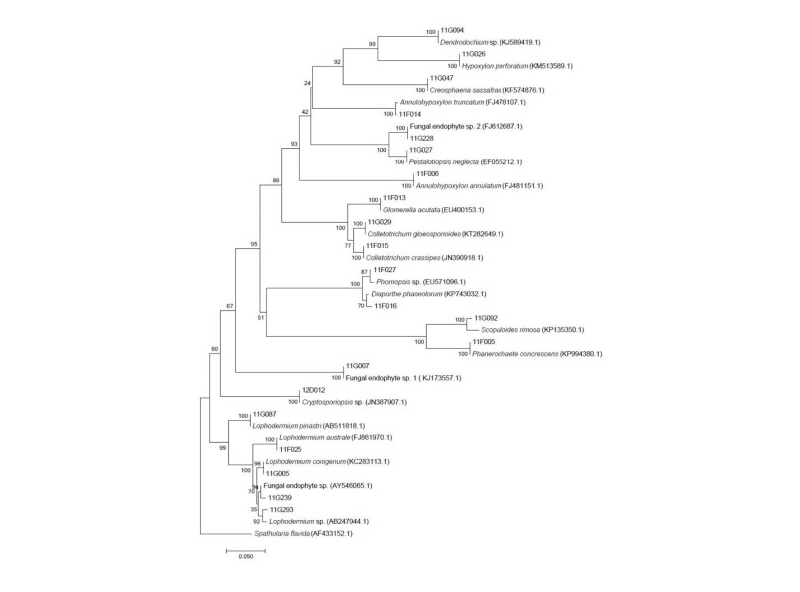
Fig. 1. Neighbor-joining phylogenic tree of endophytic fungi from Pinus densiflora and Junipreus rigida in Mt. Baekryeonsan and Johangsan. Internal transcribed spacer regions were used these sequence analysis to confirm the topological appropriation of isolates (1,000 replicates). Sphatularia flavida was used as an out-group.
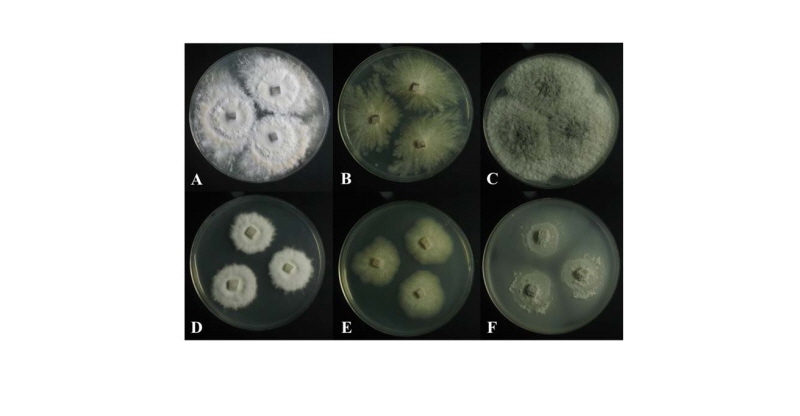
Fig. 2. Colonies of strain 11F005 (Phanerochaete concrescens) grown for 7 days on PDA (A) and MEA (D). Colonies of strain 11G047 (Creosphaeria sassafras) grown for 7 days on PDA (B) and MEA (E). Colonies of strain 11G092 (Scopuloides rimosa) grown for 7 days on PDA (C) and MEA (F). PDA, potato dextrose agar; MEA, malt extract agar.
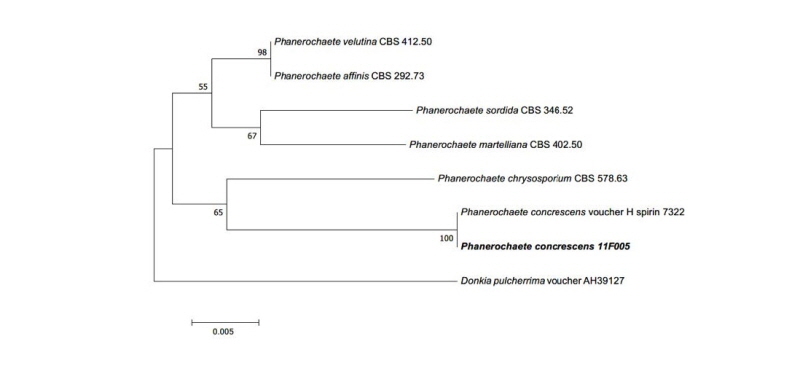
Phanerochaete concrescens V. Spirin & S. Volobuev, Mycological Progress 14:7 (2015)
Colonies on PDA measured 39~41 mm in diameter after 7 days, with a creamy color in the center, beige in the margin; colonies spreading to the border with white aerial mycelia, somewhat elevated in the center; reverse in the center, light yellow to pale brown, and ivory in the margin. Colonies on MEA measured 28~32 mm in diameter after 7 days, were restricted, with a creamy surface, somewhat elevated in the center; reverse was pale beige.
Specimen examined: Mt. Baekryeonsan, Imsil, Jeollabuk-do, Korea; N38°27′02.97′′, E126°39′08.01′′; July 22, 2011; isolated from leaves of Pinus densiflora, strain 11F005, EERSFG0000000345, GenBank no. MK158244.
Note: P. concrescens was reported from Russia in 2015 by Spirin and Volobuev. This species belonged to Basidiomycota and was isolated from Betula sp. [16]. Based on the sequence analysis, the ITS region had 99% similarity with the reference P. concrescens KP994380.1. Large subunit (LSU) also showed 99% similarity with the reference P. concrescens KP994382.1 (Fig. 3).
Creosphaeria sassafras (Schwein.) Y.M. Ju, F. San Martín & J.D. Rogers, Mycotaxon 47: 223 (1993)
Colonies on PDA measured 37~40 mm in diameter after 7 days, showed white to pale beige surfaces; flat colonies showed spreading to the border with irregular and radial shapes; reverse was dark yellow. Colonies on malt extract agar (MEA) measured 28~33 mm in diameter after 7 days, showed whitish surfaces; reverse was ivory. Colonies were restricted with irregular margins.
Specimen examined: Mt. Johangsan, Muju, Jellabuk-do, Korea; N38°27′52.00′′, E126°39′13.00′′, July 15, 2011; isolated from leaves of Juniperus rigida, strain 11G047, EERSFG0000000342, GenBank no. MK158314.
Note: C. sassafras was reported by Ju et al. in 1993 and belonged to Xylariaceae in Ascomycota [17]. In China, C. sassafras was isolated from liverwort (Scapania verrucosa) as an endophytic fungus [18]. This species can secrete antimicrobial matter as sassafrins [17]. Based on genetic analysis, the ITS region had 99% similarity with the reference C. sassafras KF574876.1. ß-tubulin (TUB) also showed 97% similarity with the reference C. sassafras KU684126.1 (Fig. 4).
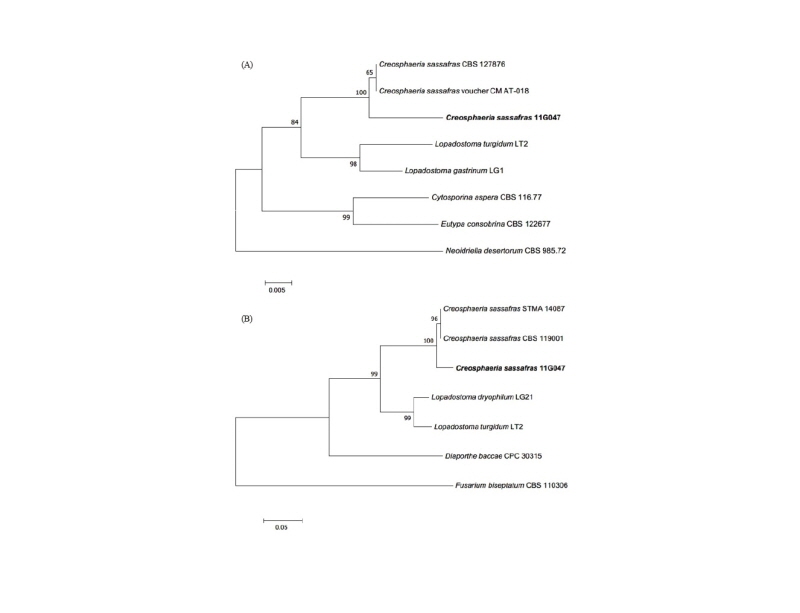
Fig. 4. Neighbor-joining phylogenetic tree based on a large subunit rDNA (A) and ß-tubulin gene (B) sequences. Neoidriella desertorum and Fusarium biseptatum were used as an outgroup in each tree. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in bold.
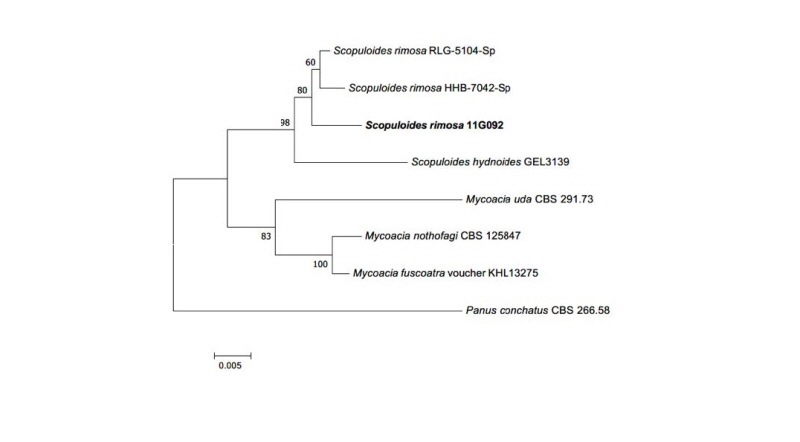
Scopuloides rimosa (Cooke) Jülich, Persoonia 11 (4): 422 (1982)
Colonies on PDA measured 43~45 mm in diameter after 7 days, with olivaceous to light gray color in the center; colonies showed spreading to the border with woolly aerial mycelia; reverse in the center was olivaceous and pale beige in the margin. Colonies on MEA measured 35~37 mm in diameter after 7 days; colonies were restricted, with slightly dense irregular mycelium in the center; the margin was complete, with a grayish green surface; the reverse was yellowish green.
Specimen examined: Mt. Baekryeonsan, Imsil, Jeollabuk-do, Korea; N38°27′02.97′′, E126°39′08.01′′, July 22, 2011; isolated from leaves of Pinus densiflora, strain 11G092, EERSFG0000000344, GenBank no. MK158344.
Note: S. rimosa was reported by Jülich in 1982 and belongs to Basidiomycota. Clampless hyphae is a specific characteristic of the genus Scopuloides [19]. Based on sequence analysis, the ITS region had 96% similarity with S. rimosa KP135350.1. LSU also showed 99% similarity with S. rimosa KP135352.1 (Fig. 5).
In Mt. Baekryeonsan, seven taxa of endophytic fungi were isolated from J. rigida, eight taxa were isolated from P. densiflora, whereas only one taxa, Annulohypoxylon turcatum, was isolated from both. In Mt. Johangsan, five fungal taxa were isolated from J. rigida, and four taxa were isolated from P. densiflora, but no common endophytic fungi were found.
Lophodermium conigenum was the most dominant endophytic fungi in P. densiflora in both mountains. A. truncatum was the dominant species isolated from J. rigida from Mt. Baekryeonsan, whereas at Mt. Johangsan, fungal endophyte sp. 2 was the dominant species.
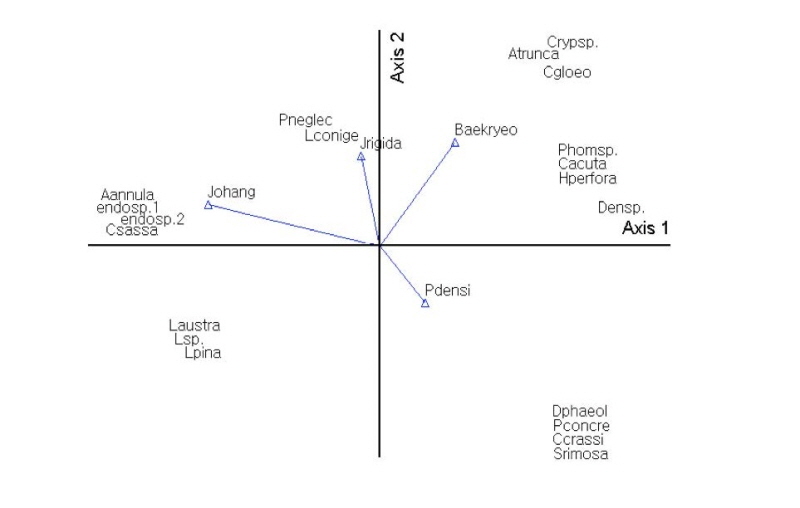
Using Bray-Curtis ordination, endophytic fungi were analyzed according to host plants and locality. The results showed that the endophytic fungal communities varied depending on host plants and locality. P. neglecta and A. annulatum were affected by host plant (J. rigida) and locality (Mt. Johangsan). Diaporthe phaeolorum and Colletotrichum gloeosporioides were affected by host plant (P. densiflora) and locality (Mt. Baekryeonsan; Fig. 6).
Lophodermium spp. were reported as pathogens of needle cast in pine trees, and approximately 103 species have been found worldwide [20]. These pathogens are known to have a unique ecological habitat according to the host plant. L. pinastri was found in senescent leaves, L. conigenum was found in conifer twigs or leaves, and L. seditiosum was found in leaves of conifer seedlings. Not all Lophodermium species showed pathogenic properties; however, some have been shown to act as endophytic fungi in conifers. In Korea, Lophodermium spp. were found to be associated with needle-cast disease [21], and six species of Lophodermium (L. austral, L. durilabrum, L. nitens, L. pinastri, L. pini-excelsae, and L. pini-pumilae) and their relationships with host plants were reported. In previous biodiversity studies of endophytic fungi in conifer trees from Korea, L. conigenum, L. pinastri, and Lophodermium sp. were found in P. densiflora, P. koraiensis, P. rigida, and P. thunbergii [3, 10, 12, 13]. Thus, Lophodermium spp. are expected to be a crucial component of fungal endophytes in host plants, and additional comparative studies of the functional roles of endophytes and conifer host plants are needed.