Introduction
The genus Dermea Fr. contains 24 species worldwide [1]. Until now only one species, D. cerasi, had been discovered in Korea on fallen branches in the national park of Byeonsanbando [2]. Since then, there was no additional record of Dermea species therein. The apothecium of Dermea is very hard, leathery, dark brown to black in color and has a clavate ascus with eight ascospores. In the asexual stage, the macroconidium is sickle-shaped or filiform with 0-3 septa, and the microconidium is rod or filiform without septa [3].
Alpine conifers are vulnerable to climate change [4]. National Institute of Ecology (NIE) has selected seven vulnerable species - Abies koreana E. H. Wilson, Abies nephrolepis (Trautv.) Maxim., Juniperus chinensis var. sargentii A. Henry, Picea jezoensis (Siebold et Zucc.) Carriere, Pinus pumila (Pall.) Regel., Taxus cuspidata Siebold et Zucc. Thuja koraiensis Nakai - and is studying their basic ecology to help conserve these species [5]. In this context, study of the biodiversity of endophytic fungi on firs is important. Through this, we have found an unrecorded endophytic fungus, Dermea piceina, on A. koreana in Korea and reported it here.
Needle leaves of A. koreana were harvested from Mt. Halla (33° 20' N, 126° 31' E, 1,952 m) in the Jeju Special Self-Governing Province of Korea in 2018. The samples were transported in a zipper bags, and fungi were isolated within 24 h. All samples were washed with tap water and cut into 1-cm pieces. Surface sterilization was performed; samples were immersed in 96% ethyl alcohol for 1 min, sodium hypochlorite for 3 min, and then 96% ethyl alcohol for 30 s; finally, they were washed twice with sterilized water. Each sample was placed on potato dextrose agar (PDA; MBcell, Seoul, Korea) and incubated in the dark for 4 weeks at 25℃ to isolate endophytic fungi [6]. PDA and maltose extract agar (MEA; MBcell, Seoul, Korea) media were used for the pure culture of endophytic fungi. The macroscopic and microscopic of the fungus were measured by light microscope (DM2500, Leica Microsystems, Wetzlar, Germany). The fungus was deposited in the Korean Collection for Type Cultures (KCTC).
Genomic DNA was extracted from the fungus using a plant tissues genomic DNA extraction kit (Xi’an Tianlong Science & Technology, Shaanxi, Taiwan) following the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed using primers ITS1 and LR3, which can selectively amplify from the internal transcribed spacer 1 (ITS1) region 1 to the D2 region of 28S ribosomal DNA [7,8]. The PCR conditions were as follows: Pre-denaturing for 5 min at 94℃ with one cycle, denaturing for 30 s at 94℃, annealing for 30 s at 50℃, extending for 1 min at 72℃ in 30 cycles, and then finally stabilizing for 10 min at 72℃ for one cycle. The PCR product was confirmed by electrophoresis using 1.5% agarose gel.
DNA sequencing was supplied to Macrogen (Seoul, Korea), and the resulting sequence was then identified based on similarity with the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the basic local alignment search tool. A maximum-likelihood tree was generated by MEGA 10.0.5 based on the Kimura-2 parameter distance model with the 1,000-times bootstrap method [9].
Dermea piceina J.W. Groves
[MB#286055]
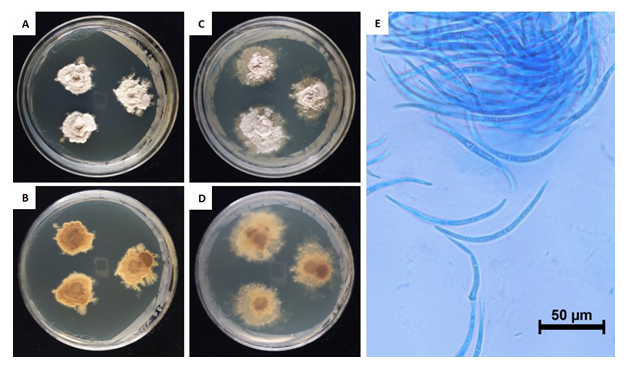
This fungus grew slowly on the chosen media. Colony diameter after 14 days was <1.0-2.0 mm on PDA and MEA. However, the colony diameter after 180 days was 15.5-19.6 mm on PDA and 21.5-30.4 mm on MEA. The mycelium was denser in PDA than in MEA. The surface color was light amberish gray (MEA) (Munsell color notation: 5Y 8/2) to light vermilionish gray (PDA) (Munsell color notation: 7.5YR 8/2) with an irregular margin, velvety texture at the center to scant at the margin, and no exudates. The reverse was grayish gamboge (MEA) (Munsell color notation: 2.5Y 7/4) to light brownish gray (PDA) (Munsell color notation: 2.5Y 9/2) [10]. Macroconidia measured 97.6-109.6×4.0-6.6 µm (n=20), were strongly curved to sickle-shaped with pointed ends, hyaline, dyed well with lactophenol cotton blue, and displayes 0-3 septa inside. Microconidia were not observed. (Fig. 1, Table 1).
Table 1. Morphological characteristics of Dermea piceina NIE7236 isolated from needle leaves of Abies korenana.
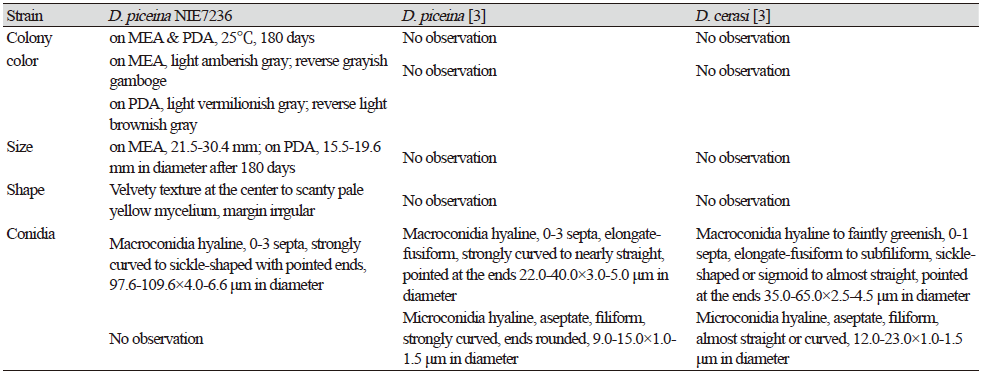
|
|
PDA, potato dextrose agar; MEA, maltose extract agar. |
Specimen examined: Mt. Halla, Jeju Special Self-Governing Province, Korea, 16 October 2018., isolated from leaves of Abies koreana, strain NIE7236, KCTC no. 56711, GenBank no. MW186178 for ITS rDNA
Note: Dermea species were identified by morphological characters (ascospore number, conidia size etc.) and host plants [3]. However, at present, host specificity is ambiguous because some studies reported their multiple host plants [11,12]. D. piceina was found on Picea sp. in its first report by Groves [3], whereas in our study, it was discovered on Abies sp.. Recently, Dermea chinensis C. M. Tian & N. Jiang, isolated from Chinese red birch (Betula albosinensis Burkill), was reported as a new species in China [1]. Most studies of Dermea spp. have been carried out in North America, Europe, and India [3] so host specificity of genus Dermea was thought to show bias due to insufficient research in other regions. The DNA sequence of D. piceina in the present study was analyzed from ITS region to the D2 region of 28S ribosomal DNA and revealed 96.8% similarity to D. piceina (MH856143.1) (Figure 2). The genus Dermea is associated with nineteen host plant genera: Abies, Acer, Amelanchier, Betula, Capparis, Chionanthus, Fraxinus, Hamamelis, Ilex, Libocedrus, Nemopanthus, Picea, Pinus, Prunus, Pseudotsuga, Rhamnus, Sorbus, Tsuga, Viburnum [1,3,12]. Through this study, we present the first report of D. piceina in Korea, and thus two species of Dermea - D. cerasi and D. piceina - are distributed in Korea. Many preceding studies of Dermea spp. focused on apothecium (ascospore) in the bark or leaf, but lack information about the morphology of colony in the media. Therefore, further studies are needed for exact measurements of their morphological characters from cultures maintained under controlled laboratory conditions.

Fig. 2.Phylogenic tree of Dermea piceina strain NIE7236 isolated from Abies koreana. The internal transcribed spacer region, including 5.8S ribosomal DNA, and 28S ribosomal DNA including D1 and D2 region, were used for the sequence analysis to confirm the topological appropriation of the fungal isolates. Diplocarpon earlianum was used as an out-group and bootstrap values are shown at the branches (1,000 replicates).