Introduction
Apple (Malus domestica) is a commercially important and widely grown crop in the temperate regions of the world [1]. It is one of the world's leading fruits, with an estimated production of 89,329,179 tons [2]. Each country and region has local own cultivars and some cultivars are familiar all over the world [3]. However, post-harvest diseases of fruits cause heavy losses during storage resulting in considerable economic losses. To date, approximately 40 pathogens were identified as the causative agents of apple diseases [4], among them apple blotch, anthracnose, white rot, Alternaria leaf spot, and bacterial shoot blight have serious economic implication for apple cultivation [5]. Several causal agents of various diseases have been identified on apple fruits such as Colletotrichum spp. [6], Botryosphaeria dothidea [7], Marssonina coronaria [8], and Fusarium decemcellulare [9]. Recently, the abnormal fruit rot symptoms were observed during the screening of post-harvest diseases on apple fruits, which were collected from orchard located in Gunwi (36°16'27.1"N 128°28'17.6"E), Korea and stored under low-temperature conditions. In the present study, the isolated fungal strains are described and illustrated as a causal agent of newly recorded post-harvest disease on apples in Korea.
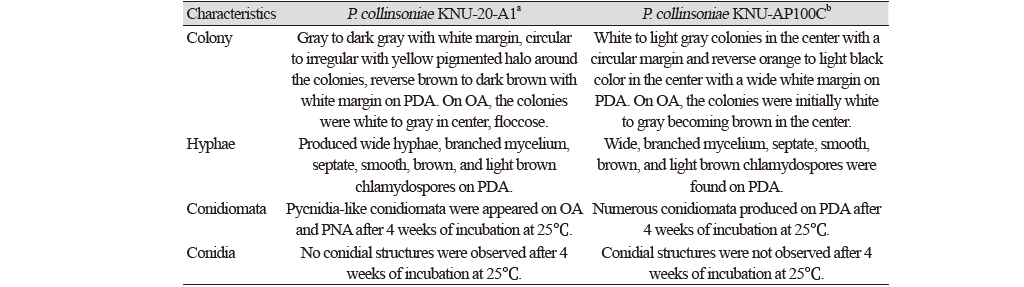
The symptoms of fruit rot disease were initially brown, later dark brown to dark red spots, irregular in shape (Fig. 1A-D), which were clearly differentiated from the typical symptoms of apple anthracnose, white rot, or Fusarium fruit rot. To isolate the causal agent from the abnormal spots, the surface of an apple was wiped with 70% ethanol and the diseased peel was removed using a sterilized blade. Then, the surface of the collected diseased tissues was sterilized for 30 seconds in 70% ethanol and 1% sodium hypochlorite and washed three times with sterilized double-distilled water. The surface-sterilized tissues were transferred onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and maintained in an incubator at 25℃ [10]. As the result, the three strains were isolated from the diseased apples and then designated as KNU-20-A1, KNU-20-A2, and KNU-20-A3. To analyze the cultural characters, PDA, oatmeal agar (OA; Difco, Detroit, MI, USA) and autoclaved pine needle on 2% water agar (PNA) were prepared for the detail description of the isolated fungal strains [11]. After 10 days of incubation on PDA at 25℃, the colonies were 23-25 mm in diameter, gray to dark gray with white margin, circular to irregular with yellow pigmented halo around the colonies, reverse brown to dark brown with white margin (Fig. 1E and F). On OA, the colonies were 36-38 mm in diameter, white to gray in center, floccose, reverse brown with yellow margin (Fig. 1G and H). After 4 weeks of incubation, pycnidia-like conidiomata structures were appeared on OA and PNA, round to irregular, solitary to aggregate, and dark brown to black, and with the diam. of 600-750 μm (Fig. 1M and N). However, no conidial structures were observed. The strains also produced wide hyphae, branched mycelium, septate, smooth, brown, and light brown chlamydospores on PDA (Fig. 1O and P). The cultural and morphological characteristics were found to be similar with those of previously identified Plenodomus collinosinae (Table 1) [12]. The type strain of Leptosphaeria collinsoniae (=Plenodomus collinsoniae) has been reported as a sexual morph on the host called Collinsonia canadensis, and after that the strain was also combined with the strain P. collinsoniae CBS 120227 isolated from Vitis coignetiae, with the Perithecia; scattered, gradually blackening the stems, covered by the cuticle, finally bare, globose-conic, rugose, papillate, Asci; terete, short-stipitate, Sporidia; amber colored, biseriate, 5-8-septate, mostly 6-nucleate, and the asexual morph was not determined [13].

Fig. 1.Disease symptoms, cultural and morphological characteristics, and pathogenicity test on apples (cv. Fuji) with inoculation using colony agar blocks of Plenodomus collinsoniae KNU-20-A1. (A-D): Primary symptoms; (E, F): Colony of strain KNU-20-A1 on potato dextrose agar (PDA) after 10 days; (G,H): Colony of strain KNU-20-A1 on oatmeal agar (OA) after 10 days; (I, J): Inoculation after 14 days; (K, L): Enlarged picture; M: After 30 days, pycnidia-like structures on PNA (pine needle agar); (N): After 30 days, pycnidia-like structures on OA agar; (O, P): Hyphal structures of P. collinsoniae KNU-20-A1 (scale bars: M, N=500 μm; O, P=10 μm).
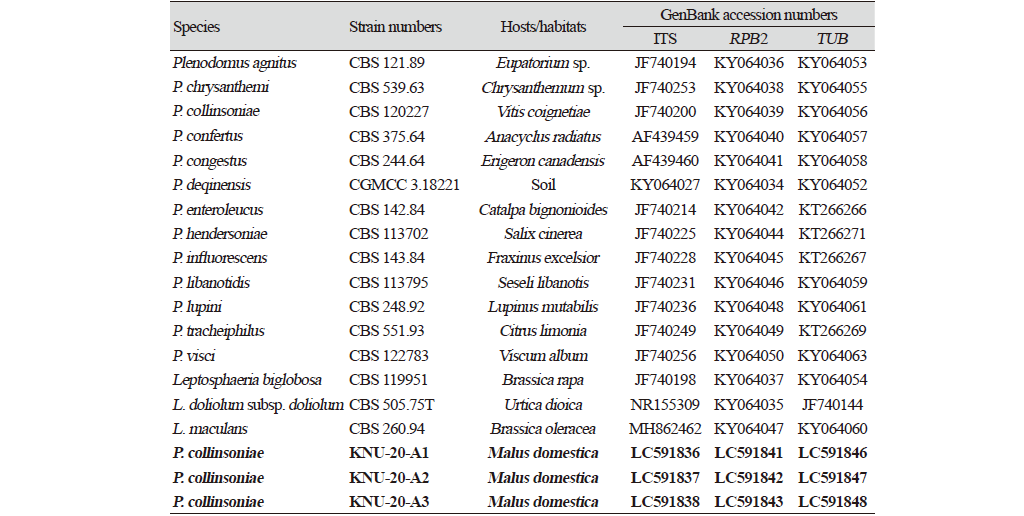
For molecular identification, the total genomic DNA was extracted from the three above-mentioned strains using a HiGene Genomic DNA prep kit (BIOFACT, Daejeon, Korea) following the manufacturer's instructions. The ITS regions, beta-tubulin (TUB), and the second largest subunit of RNA polymerase II (RPB2) genes were amplified using the primer sets ITS1F/ITS4 [14,15], Btub2Fd/Btub4Rd [16], and RPB2-5F2/fRPB2-7cR [17], respectively. Amplified PCR products were purified with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Macrogen Co., Ltd. (Daejeon, Korea). From the sequence analysis, sequences from the strains KNU-20-A1 (525, 344, and 1062 bp), KNU-20-A2 (567, 346, and 1046 bp), and KNU-20-A3 (553, 328, and 1035 bp) were obtained from the ITS regions, TUB, and RPB2 genes, accordingly. Comparative sequence analyses of the molecular markers of the three strains revealed high similarities of 99.8-100%, indicating their affiliation to the same species. A BLAST search of the NCBI database using sequences of ITS regions revealed the highest similarity of strains KNU-20-A1, KNU-20-A2, and KNU-20-A3 with Plenodomus collinsoniae CBS 120227 (99.6%). And the closest species P. influorescens CBS 143.84 and P. visci CPC 35316 showed maximum 92.1-92.5% and 90.4%, respectively from the ITS regions. The partial sequences of RPB2 gene of the three strains shared maximum 98.7% identity with that of P. collinsoniae CBS 120227, and only 89.4% and 90.7% similarities with the other closest relative, P. influorescens CBS 143.84 and P. visci CBS 122783, respectively. Based on the TUB gene sequence the three above-mentioned strains were close to P. collinsoniae CBS 120227 (97.9-98.2%), P. influorescens CBS 143.84 (85.4-86.1%), and P. visci CBS 122783 (86.8-87.5%). Using the three molecular markers, ITS regions, RPB2 and TUB genes, the closest neighbor of the three isolated strains were determined to be P. collinsoniae with the high values of the sequence similarities of 97.9-99.6% while no difference in sequences containing more or less base pairs between the markers from the isolated fungal strains. To confirm the relationship of the above-mentioned strains with P. collinsoniae at the species level, phylogenetic analysis using concatenated sequences of the ITS regions, TUB and RPB2 genes were performed. The sequences of allied species were retrieved from the National Center for Biotechnology Information (NCBI) (Table 2). Phylogenetic trees were constructed using neighbor-joining (NJ) [18], maximum-likelihood (ML) [19], and maximum-parsimony (MP) [20] methods, as implemented in MEGA7.0 [21]. The alignments were performed for each gene, and then the sequences were merged by using MEGA7.0 software program. The NJ analysis was performed using Kimura two-parameter distances [22] with gaps excluded from the analysis. A bootstrap analysis with 1000 replicate was performed to assess the support for clusters. In the phylogenetic tree (Fig. 2) all isolated strains occupied a position within the genus Plenodomus and clustered together with P. collinsoniae, indicating their closest relationship at the species level. Thus, strains KNU-20-A1, KNU-20-A2, and KNU-20-A3 were identified as P. collinosinae based on multi-locus phylogenetic analysis along with their cultural and morphological characteristics, which were completely consistent with those previously reported for this fungal species [12,13].

Fig. 2.Neighbor-joining phylogenetic tree based on the concatenated sequences of the internal transcribed spacer (ITS) regions, beta-tubulin (TUB), and RNA polymerase II (RPB2) genes showing the phylogenetic position of the three isolated strains (KNU-20-A1, KNU-20-A2, KNU-20-A3) among Plenodomus species and other closely related taxa. Bootstrap values (based on 1000 replications) greater than 50% are shown at branch points. The isolated strains are shown in bold. Bar, 0.02 substitutions per nucleotide position.
To confirm the pathogenicity of P. collinosinae isolated in this study, KNU-20-A1 was selected as the representative from the three strains and inoculated into healthy apples (cv. Fuji) fruits with three replications. The inoculum was prepared using strain KNU-20-A1 cultured for 4 weeks on PDA. The surface of a healthy apple was wiped with 70% EtOH and then air-dried. Two points of apple were wounded using a sterilized needle and colony agar blocks were attached and sealed using foil. Apple fruits inoculated with sterilized water were used as the control. All the inoculated fruits were incubated at 25℃, and after 3 days the colony agar blocks were removed. After 14 days, brown with slightly sunken spots were observed on apples which were identical to typical primary symptoms (Fig. 1I-L) while no symptoms were observed in unwounded fruits (data not shown). From each of the inoculated fruits P. collinosinae was re-isolated and the cultural and morphological characteristics were compared with those of the inoculated strains, and all characteristics were found to be the same (Table 1).
In a previous study, the members of the Plenodomus seem to be cosmopolitan in distribution, since they have been recorded from both temperate and tropical countries (i.e. China, Greece, France, Japan, Netherlands, Peru, Spain, Taiwan) [23]. Until recent, 100 epithets of Plenodomus have been listed in the Index Fungorum database [24]. The host specificity of Plenodomus has not yet been clarified based on species from different plant families (Asteraceae, Lamiaceae, Liliaceae) [13]. P. meliloti was a low-temperature parasitic fungus found only in the provinces of Alberta and Saskatchewan in Canada [25], whereas, P. morganjonesii was obtained from partially degraded leaves from New Jersey [26]. P. chrysanthemi was isolated as an endophyte of Chenopodium album that represents a new host from Iran [27]. The causal agent of foot rot and storage tuber rot on sweet potato was identified as P. destruens from experimental fields in China [28]. A novel species, P. sinensis was introduced from Tamarindus indica L. (Fabaceae) and Plukenetia volubilis L. (Euphorbiaceae) in Yunnan Province, China [29]. Moreover, Brown root rot, caused by the fungal pathogen Phoma sclerotioides (=Plenodomus meliloti), was associated with winterkill, slow emergence from winter dormancy, and yield loss of alfalfa (Medicago sativa L.), and also was a problem with severe winters in Alaska and Alberta, Saskatchewan and Manitoba, Canada [30]. Although many fungal species belonging to the genus Plenodomus were isolated in several countries from diversified hosts, only P. destruens was reported as causing agent of storage tuber rot of sweet potato in Korea [31], but there are no studies of P. collinsoniae related to plant diseases in Korea. Recently, there are two species of Plenodomus, namely P. sinensis and P. collinsoniae that were isolated from a soil sample collected in abandoned apple orchard in Gyeongsangbuk-do, Korea [12]. Even, there is no enough information that the genus Plenodomus can cause diseases in apples, but there might be a relation or transfer of causal agents from soil to apples trees or fruits as well as causing disease. Furthermore, the genus Plenodomus includes several well-known important plant pathogens and is found as opportunistic fungi on several hosts. In this study, the strains were isolated from the infected disease apples (cv. Fuji) fruit and identified the fungal pathogen caused post-harvest disease on apples in Korea.
Furthermore, based on the results of pathogenicity test, the disease symptoms appeared very slowly on the inoculated apple fruits, so it is assumed that it can be observed in the long-term stored period in the low-temperature storage room condition. Still, there were no reported apple diseases caused by Plenodomus species in Korea. According to the results of the present study, P. collinsoniae can be a new fungal agent of the post-harvest disease of apple, and the ecology of P. collinsoniae should be further studied for the proper control. In conclusion, this is the first report of post-harvest disease on apple caused by P. collinsoniae in Korea.