Introduction
Environmental changes and mutations in specific genes can lead to the emergence of mycoviral infections [1]. Viral infections in mushrooms are caused by mycelium fusion, which also transmits the virus to healthy mycelium by spores [2]. Similar to mushroom spores, mycelium remaining in the mushroom cultivator can be a viral infection medium [3].
Lentinula edodes is a commercially important, edible mushroom that is cultivated throughout Korea. Compared to Flammulina velutipes and Pleurotus ostratus, L. edodes has a relatively long cultivation period, with fruiting bodies that can be harvested more than five times on the same growth medium. The cultivation of L. edodes in farms is often exposed to the surrounding environment, which can increase the risk of viral infections [4,5].
Since the first description of fungal viruses 50 years ago, there has been an increase in the number of mycoviruses that have been identified [6]. Viral infections in L. edodes was first reported in Japan during the 1970s [7], and viruses associated with L. edodes have also been found in Korea in more recent studies [4,8]. LeV-HKB was the first mycovirus that was identified to have a dsRNA genome, and it shares a high degree of similarity with the genomes of LeSV and LeV-HKB [8,10]. LePV1 and LeV-HKB can coinfect L. edodes, and studies have also been performed to evaluate how these infections affect each other [9].
It is essential to screen strains that are not infected with a virus as viral infections have a significant impact on mushroom quality and productivity [11]. Since infection of the mycovirus is difficult to eradicate and it is necessary to use virus-free strains to prevent damage caused by the mycovirus [12,13].
The cultivation conditions of some mushrooms can help to prevent the impact of viral infections, even if they are strains that contain viral particles [14-16]. If viral particles are found but no disease appears, the viral infection may be overlooked; however, the same strain can cause problems under different conditions. Therefore, viral detection is strains used for breeding is paramount to the successful cultivation of mushrooms.
In this study, we use reverse transcription-polymerase chain reaction (RT-PCR) to detect RNA mycoviral infection in wild strains of L. edodes that can be used as a breeding resource in the future. The presence of five RNA mycoviruses (LePV1, LeSV, LeV-HKB, LeNSRV1, and LeNSRV2) were tested in the wild strains of L. edodes, and we observed the occurrence of single and multiple viral infections within a single wild strain.
Materials and methods
Fungal strains
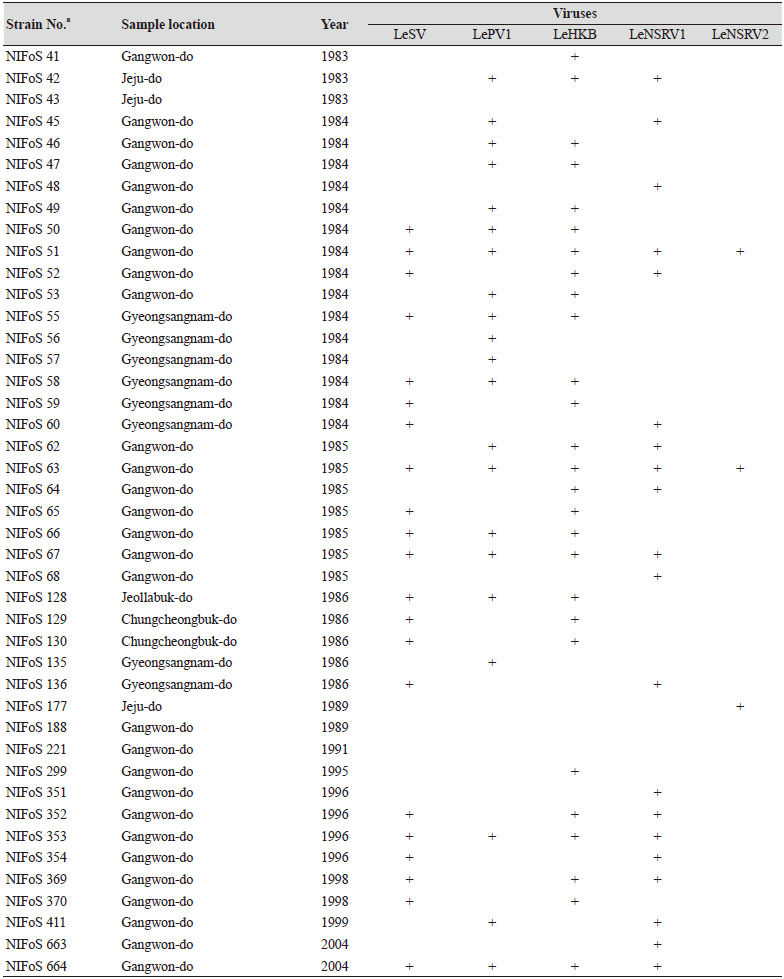
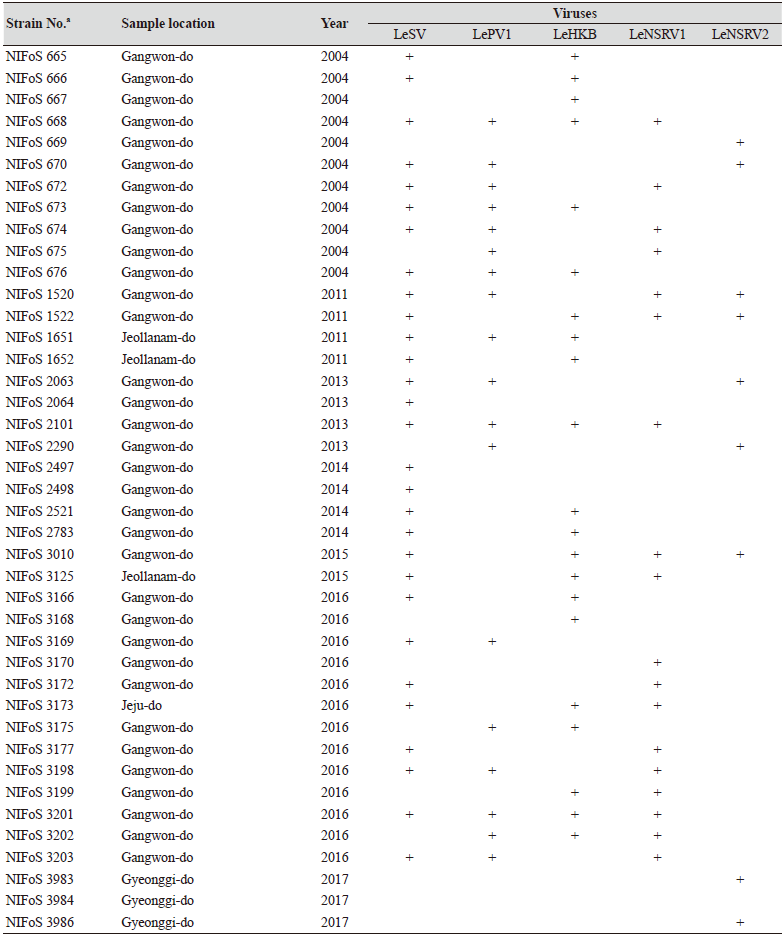
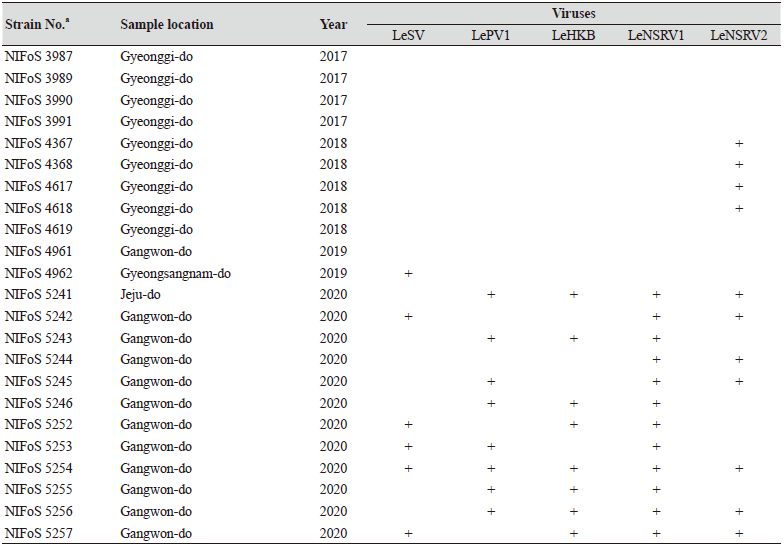
One hundred and twelve wild strains of L. edodes were examined in this study (Table 1). They were isolated from samples that were collected from various locations across Korea from 1983 to 2020 and maintained at 4℃ through successive subcultures by the National Institute of Forest Science. The strains were cultured on potato dextrose agar (PDA, Difco, MD, USA) media at 25℃ for two weeks under dark conditions before use.
Table 1. The sampling information and RT-PCR analysis of the occurrence of RNA mycoviruses in wild strains of Lentinula
edodes in Korea.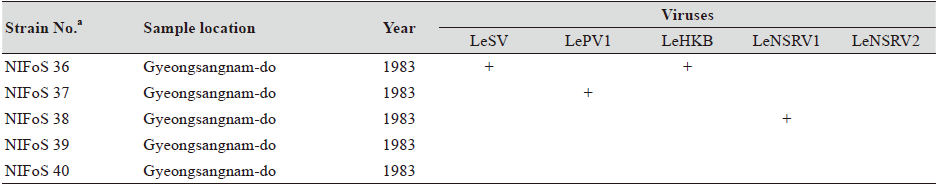
|
RNA extraction and cDNA synthesis
The mycelium grown on each PDA medium was collected by a cell scraper and frozen in liquid nitrogen. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, CA, USA), following the manufacturer’s instructions. The quantity and quality of the RNA were examined using an Epoch Multi-volume Spectrophotometer (BioTek, VT, USA). Single strand cDNA synthesis was carried out using Moloney murine leukemia virus reverse transcriptase (New England Biolabs).
RT-PCR
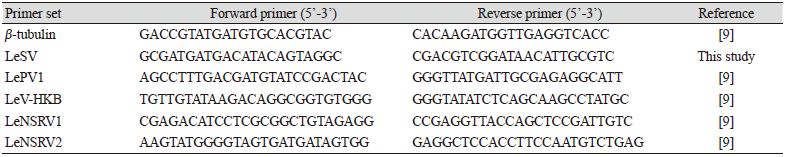
To confirm viral infection, RT-PCR was conducted with specific primers to amplify the RNA-dependent RNA polymerase (RdRp) genes of each mycovirus (Table 2). Amplification of the β-tubulin gene was used as a positive control to confirm successful RNA extraction and cDNA synthesis. All the primers used for RT-PCR are listed in Table 2. RT-PCR for β-tubulin and LeSV were performed under the following conditions: 95℃ for 5 min; 25 cycles of 95℃ for 30 s, 55℃ for 30 s, and 72℃ for 1 min; 72℃ for 5 min. RT-PCR for LePV1, LeV-HKB, LeNSRV1, and LeNSRV2 were performed under the following conditions: 95℃ for 5 min; 35 cycles of 95℃ for 30 s, 60℃ for 30 s, and 72℃ for 1 min; 72℃ for 5 min. The PCR products were confirmed using 1.5% (w/v) agarose gel electrophoresis.
Results and Discussion
We isolated 112 wild strains of L. edodes from different times and locations in Korea (Table 1); there were 75 strains from Gangwon province, 14 strains from Gyeongsangnam province, 12 strains from Gyeonggi province, 5 strains from Jeju province, 3 strains from Jeollanam province, 2 strains from Chungcheongbuk province, and 1 strain from Jeollabuk province.
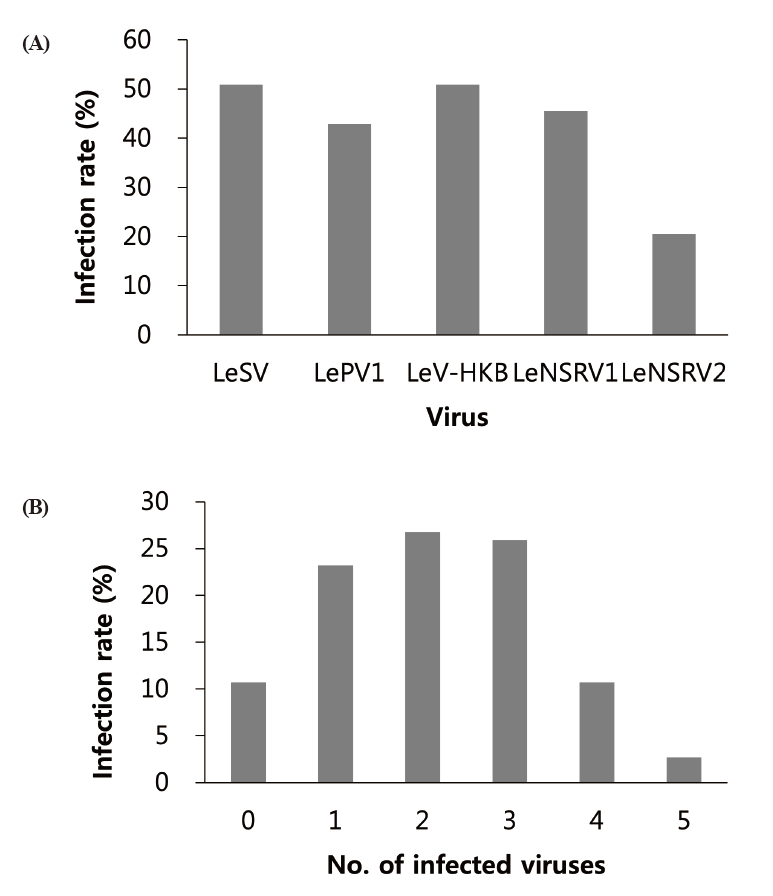
Specific primer sets (LeSV, LePV1, LeV-HKB, LeNSRV1, and LeNSRV2) were used (Table 2) to detect viral infection. According to the RT-PCR analysis, products of 328 to 394 bp in length were amplified from the wild strains when infected with the viruses (LeSV 386 bp, LePV1 389 bp, LeV-HKB 394 bp, LeNSRV1 377 bp, and LeNSRV2 328 bp) (Fig. 1).
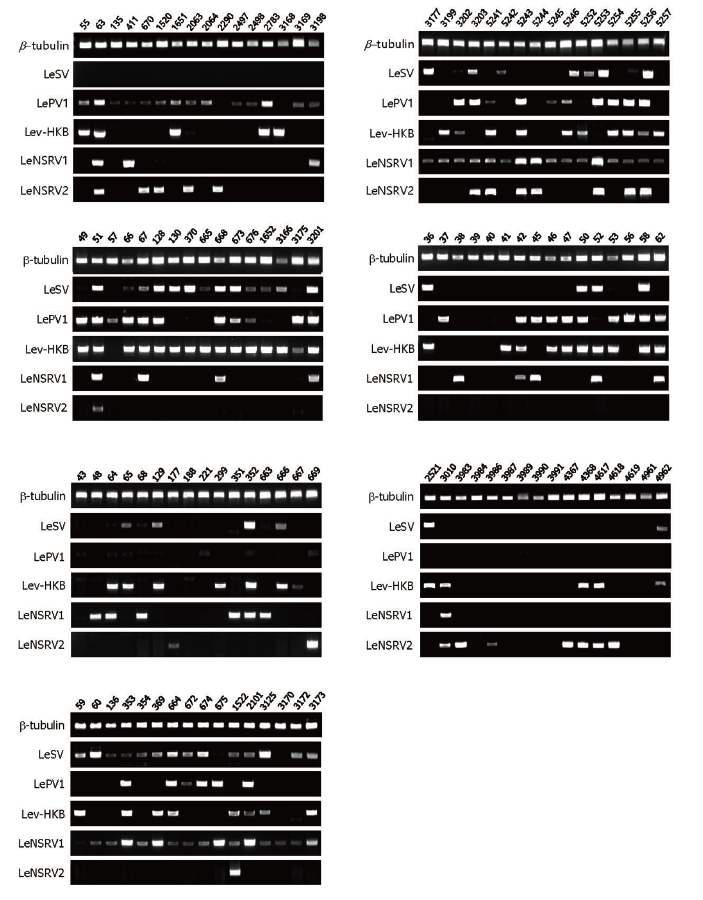
Fig. 1. Reverse transcription-polymerase chain reaction (RT-PCR) detection of viral infection from 112 wild strains of Lentinula edodes . Specific primer sets targeting RNAdependent RNA polymerase genes of each RNA mycovirus were adopted for RT-PCR. Numbers represent each of the wild strains. Amplification of the β -tubulin gene was used as the positive control to confirm the RNA extraction.
Approximately half of the strains were infected with LeSV (50.9%, 57/112), LeV-HKB (50.9%, 57/112), LeNSRV1 (45.5%, 51/112), or LePV1 (42.9%, 48/112) viruses. The LeNSRV2 virus had a lesser infection than the other viruses (20.5%, 23/112) (Fig. 2A).
Most of the infected strains were co-infected with different viruses (Fig. 2B). Among them, 30 strains (26.8%) were infected with two different viruses and 29 strains (25.9%) were infected with three different viruses. One viral infection was observed in 26 (23.2%) strains. Only three strains, NIFoS 51, 63, and 5254, that were collected from Gangwon province had five mycoviruses. On the other hand, 12 strains (10.7% of the total wild strains) were not infected by the viruses.
In this study, we observed multiple infections in which two or more types of virus were present in a single strain. Although multiple viral infections are often found in plants and fungi, much research is still needed on the consequences of multiple infections [17,18]. For example, the African cassava mosaic virus (ACMV) and East African cassava mosaic virus (EACMV) are commonly found to co-infect cassava plants with cassava mosaic disease (CMD) symptoms [19]; viral symptoms were more severe when ACMV and EACMV were present in Nicotiana benthamiana plants [19]. These results indicate that the two viruses have synergistic interactions [19]. The single infection of sweet potato feathery mottle virus (SPFMV, genus Potyvirus) and sweet potato chlorotic stunt virus (SPCSV, genus Crinivirus) in sweet potato do not show significant symptoms [20]. In contrast, coinfection with the two viruses is caused by the sweet potato virus disease (SPVD), with severe symptoms evident in plants [20]. With the discovery of LePV1 and LeV-HKB coinfection in L. edodes, studies have also been conducted to assess their influence on each other [9]. When the LePV1 virus is coinfected with LeV-HKB, the RdRp gene expression has been reported to be higher than that of a single infection [9].
Furthermore, how multiple infections of the virus found in L. edodes affect a viral disease should be further studied. Despite the difficulty in evaluating the effects of each virus when several viruses infect one strain, a virus-cured strain can be used as an important control to investigate the individual effects. It is also necessary to further analyze whether a virus is present in the fruiting body by single spore isolation in the event of hybridization.
Although the number of strains collected from provinces other than Gangwon province was small, it is interesting to note that geographically LeSV, LePV1, LeV-HKB, and LeNSRV1 were not present in the strains collected from Gyeongsangnam province. Further, only the LeNSRV2 virus was detected in the samples collected from Gyeonggi province. These phenomena suggest the possibility that the infection of each virus is related to regional characteristics. To increase the diversity of wild strains of L. edodes in the future, we must investigate forest areas throughout the country, and the confirmation of viral infection in these collected strains will be essential.