Fungal endophytes are fungi that inhabit the tissues of healthy plants without any disease symptom [1]. Leaves of evergreen woody plants have a wider parenchyma and intercellular space area per unit area compared with that of leaves from deciduous trees [2]. Endophytes inhibit plant pathogens of evergreens through production of ethyl acetate or antifungal metabolites [3]. Endophytes found in evergreen trees differ from those found in deciduous trees in terms of species composition and diversity [4]. In this study, two endophytic fungal species, Muyocopron lithocarpi and Didymocyrtis cladoniicola were isolated from the healthy leaves of Ligustrum japonicum Thunb. and Chamaecyparis obtuse (Siebold & Zucc.) Endl, two evergreen woody plants native to Hansando Island. The fungal species were discovered while studying the biodiversity of endophytic fungi in evergreen woody plants; they have not previously been recorded in South Korea. Based on the molecular and morphological characteristics, the species were confirmed by comparison with the first reported descriptions.
The leaves of L. japonicum and C. obtuse used in the study were collected from Hansando Island (N34° 46′ 77″, E128° 29′ 05″) in April, 2019. Samples were transported to the laboratory using polyethylene bags and separated within 24 h. For surface sterilization of leaves, the surface was washed with distilled water and 1% NaClO solution for 1 min, and then with 70% ethanol for 2 min [5]. The sterilized leaves were cut into 1.5–2 cm length and placed on potato dextrose agar (PDA) medium. The mycelium extending from the leaf was cut with a knife and repeatedly transferred to PDA medium to isolate it [6]. The fungal isolates retained were cultured in PDA medium and malt extract agar (MEA) medium at 25°C for 7 d. Genomic DNA was extracted according to the manufacturer’s protocol for the DNeasy plant mini kit (Qiagen, Germantown, MD, USA). The extracted DNA was subjected to polymerase chain reaction (PCR) using the primer pairs ITS1F/ ITS4 and LR0R/LR16 [7, 8]. Each primer pair was used to amplify the internal transcribed spacer (ITS) region and the large subunit (LSU) region of ribosomal DNA, respectively. ITS and LSU sequences were identified based on their similarity with the sequences recorded in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the basic local alignment search tool (BLAST). By combining the ITS and LSU regions, a neighbor-joining tree was created using the MEGA 7.0.26 program [9]. The analysis model used the Kimura-2 parametric distance model, and reliability was evaluated with the 1,000-times bootstrap method.
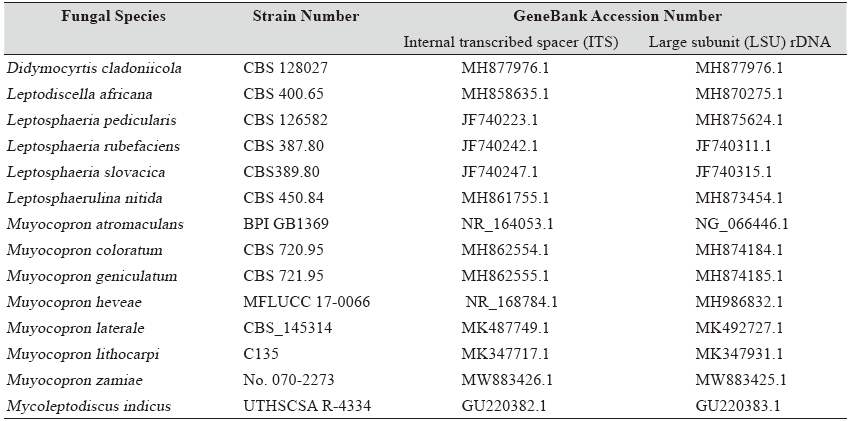
Table 1. Morphological characteristics of endophytic fungal strains isolated in this study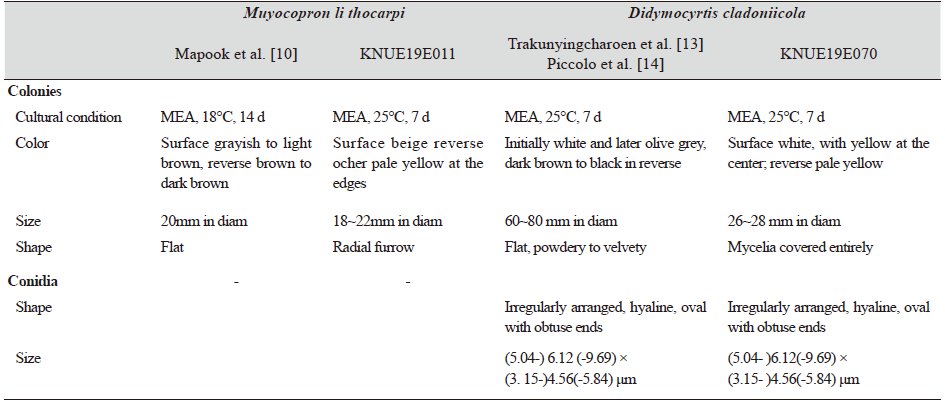
|
|
MEA, malt extract agar |
Muyocopron lithocarpi Boonmee & K.D. Hyde, Phytotaxa 265 (3): 235 (2016) [MB#551618]
Morphological Characteristics of Strain KNUE19E011. The size of the colony cultured in PDA medium for 7 days was 22–24 mm. The surface of the colony was grayish white in PDA, with an irregular margin and a texture similar to down or cotton (Fig. 1). The reverse surface was yellow, and the edges were pale yellow. After 7 days of culture in MEA medium, the size of the colony was 18–22 mm, the center of the top surface was beige, and the edges were white. Irregular radial furrows appeared on the surface, which was higher than the surface of the medium. The center of the reverse surface was ocher, and the edges were pale yellow (Fig. 1).
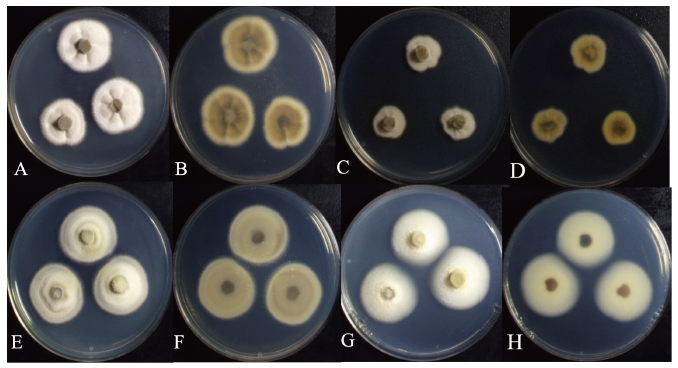
Fig. 1. Cultural characteristics of two endophytic fungal strains. Colonies of Muyocopron lithocarpi KNUE19E011 grown for 7 days on potato dextrose agar (PDA) (A: Surface, B: Reverse) and malt extract agar (MEA) (C: Surface, D: Reverse); Colonies of Didymocyrtis cladoniicola 19E070 grown for 7 days on PDA (E: Surface, F: Reverse) and MEA (G: Surface, H: Reverse).
Specimen examined. Hansando Island, Tongyoung-si, Korea, N34° 46′ 9.41′′, E128° 30′ 24.99′′, April 5, 2019. M. lithocarpi KNUE19E011, isolated from needles of L. japonicum, NIBRFG0000506585, GenBank No. MZ206175 (ITS), MZ254891 (LSU).
Note. Muyocopron lithocarpi was first discovered on Lithocarpus lucidus in Chiang Rai, Thailand in 2016. Its name was derived from the host from which the fungus was first isolated [10]. Although the morphological characteristics of asci and ascospores of M. lithocarpi were similar to those of M. umbilicatum, they were considered as different species owing to the average size of their ascospores. [10]. The KNUE19E011 strain isolated in this study was confirmed as M. lithocarpi by comparing its morphological characteristics with the Boonmee and Hyde’s description and phylogenetic analysis. BLAST result showed 100.0% similarity with M. lithocarpi MK347717.1 for the ITS region and 99.67% similarity with M. lithocarpi KU726967.1, for LSU. Phylogenetic tree analysis showed that strain 19E001 clustered together with M. lithocarpi C135 showing 99% bootstrap values (Fig. 2, Table 2).
Didymocyrtis cladoniicola (Diederich, Kocourk. & Etayo) Ertz & Diederich [MB#814023] [MB#810830]
Morphological Characteristics of Strain KNUE19E070. The size of the colony cultured in PDA medium for 7 days was 22–25 mm, the top surface was beige, and the center and edges were light olive. The back of the PDA medium was olive-colored, and the edges were pale yellow. The surface of the colony has a texture similar to down, and the edges were smooth and round (Fig. 1B). The size of the colony cultured for 7 days in MEA medium was 26–28 mm. The top of the MEA medium was white, and the center was light yellow. As in PDA medium, the surface of the colony was covered with mycelium with a texture similar to that of down. The conidia were smooth, oval, and colorless or light brown. The diameter was 2.86 (1.54–3.72) × 5.38 (3.24–6.59) μm.
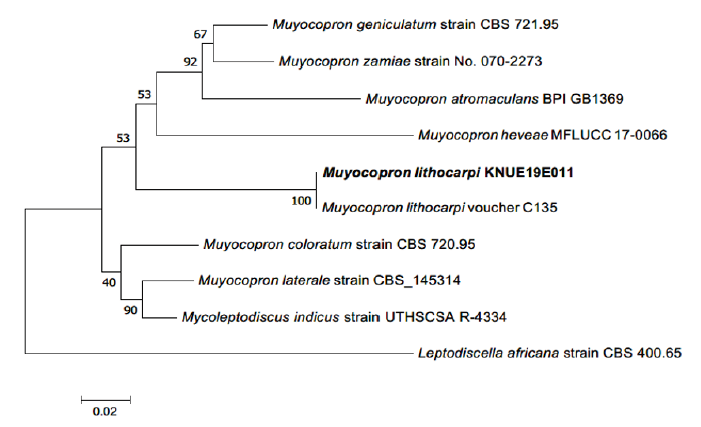
Fig. 2. Neighbor-joining phylogenetic tree based on a concatenated alignment of internal transcribed spacer (ITS) and large subunit (LSU) rDNA sequences. Leptodiscella africana was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). The fungal strain isolated in this study is in bold.
Specimen examined. Hansando Island, Tongyoung-si, Gyeongsangnam-do, Korea, N34° 47′ 57.70′′, E128° 28′ 27.55′′, April 5, 2019. D. cladoniicola KNUE19E070, isolated from needles of Chamaecyparis obtusa, NIBRFG0000506594, GenBank No. MZ206178 (ITS), MZ221116 (LSU).
Notes. Didymocyrtis cladoniicola was first discovered on the thallus of Cladonia pyxidata, a lichen species, in USA [11]. D. cladoniicola is generally found in lichens, however, the species isolated in this study had been isolated as a leaf endophytic fungi from Geum peckii, a herbaceous plant belonging to the Rosaceae [12]. BLAST results showed a 98.47% similarity with D. cladoniicola LT796877.1 for the ITS region, and a 99.67% similarity with D. cladoniicola LN907456.1 for the LSU region. Phylogenetic tree analysis showed that strain 19E070 clustered together with D.cladoniicola CBS 128023 and CBS 128025 showing 94% bootstrap values (Fig. 3, Table 2).
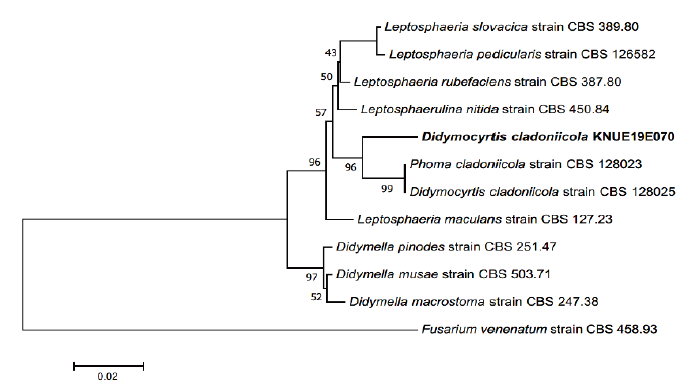
Fig. 3. Neighbor-joining phylogenetic tree based on a concatenated alignment of internal transcribed spacer (ITS) and large subunit (LSU) rDNA sequences. Fusarium venenatum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). The fungal strain isolated in this study are in bold.