Introduction
Freshwater ecosystems have many biological resources and play an important ecological role in the material cycle, energy flow, and environmental maintenance [1]. Biological resources are vital for the development of healthy freshwater ecosystems and have significant economic and social value [,]. Among these diverse biological resources, microorganisms are promising materials with great potential for use in the food, medicine, and biomass energy fields [3,4]. In particular, fungal resources are prospective materials for the development of biocontrol agents, medicines, cosmetics, and food [5,6].
Many recent studies have indicated that fungi are abundant in freshwater ecosystems. With advances in DNA sequencing technology, diverse fungal taxa have been discovered in a range of freshwater habitats [7-9]. Culture-independent detection provides information on the taxonomic diversity of aquatic fungi [10-12]. However, studies on freshwater fungi to determine their ecological functions and physiological features are still lacking. To understand the physiological ecology of fungi, cultivation followed by isolation from their natural habitats is necessary. Cultivation-based techniques facilitate the identification of morphological, physiological, and biochemical characteristics of fungi for subsequent use as biological resources.
According to previous studies, the predominant fungal genera in freshwater habitats are Aspergillus, Penicillium, and Cladosporium [13-16], which have been frequently isolated from freshwater using culture-based methods. However, in the present study, three fungal species that are not linked to the predominant genera were isolated from reservoirs in Korea, namely Paraphoma radicina, Sordaria macrospora, and Mortierella biramosa. These have not been previously reported in Korea. We investigated morphological and molecular phylogenetic characteristics of these fungal species.
Materials and Methods
Fungal isolation
Soil sediment, clams, and freshwater samples were collected from the reservoirs (Table 1). To isolate fungal strains from the soil sediment, a dilution plate method was used with a 1:10 dilution. The soil suspension was spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA). For the water samples, a simple plating technique was used for spreading the samples onto the PDA. The clamshells were washed with distilled water and cut into 2 mm2 pieces, before being placed on the PDA. Rifampicin (15 ppm) was added to the medium to limit bacterial growth. After incubation for 2-3 days at 25℃ in the dark, new hyphal tips were isolated from the outgrowing mycelia and then transferred onto fresh PDA. The new isolates were incubated for approximately 10 days at 25℃. The resultant cultures were deposited at the Nakdonggang National Institute of Biological Resources (NNIBR).
Table 1. Information on Mortierella , Paraphoma , and Sordaria isolates from reservoirs in Korea.
|
| ITS: the internal transcribed spaer of rDNA. |
Cultural and morphological analyses
The characteristics of the cultures were investigated 3-7 days after inoculation of the isolates on PDA at 25℃ in the dark. After a week, the microscopic structures of the isolates were observed under a Zeiss Axio Imager A2 microscope (Carl Zeiss, Oberkochen, Germany), and then photographed using an Axiocam 512 color camera (Carl Zeiss).
DNA extraction, amplification, and sequencing
The genomic DNA of the strains was isolated using the MagListo 5 M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea), based on magnetic bead technology. The 28S large subunit (LSU) and internal transcribed spacer (ITS) of ribosomal DNA regions were amplified with PCR using primer sets LROR/LR5 [17] and ITS1/ITS4 [18], respectively. The partial nuclear β-tubulin gene of the Sordaria isolate was amplified using Bt2a/Bt2b [19]. The DNA amplicons were purified using an AccuPrep PCR Purification Kit (Bioneer), and sequenced by Macrogen, Inc. (Seoul, Korea). The sequences were registered in the National Center for Biotechnology Information (NCBI) GenBank database (Table 1).
Phylogenetic analysis
The DNAStar software package version 5.05 (DNAStar, Inc., Madison, WI, USA) was used for sequence editing. The edited sequences were blasted to search for the closest reference sequences from the NCBI. The sequence data for Korean strains and reference sequences from previously published type or authentic isolates were aligned using MAFFT 7 [20], with the Q-INS-i algorithm [21]. Minimum evolution (ME) and maximum likelihood (ML) analyses were used to construct phylogenetic trees with MEGA X [22] using the Tamura-Nei model. Bootstrapping analysis was performed with 1,000 replicates. Multilocus phylogenetic analysis was performed after the sequences of the individual markers had been concatenated in SequenceMatrix v1.7.8 [23].
Results
The three fungal strains W251, W471, and W913, were isolated from water, shellfish, and soil sediment, respectively, from three different reservoirs, and were investigated phylogenetically and morphologically.
The isolate W251 belonging to the species Paraphoma radicina matched the epitype strain of CBS 11179 (NR_156556 in ITS, NG_070446 in 28S rDNA) with sequence similarities of 100% (486/486 bp) in ITS and 99.8% (843/845 bp) in 28S rDNA in the BLASTn search. The ITS and 28S rDNA sequences of isolate W471 belonging to the Sordaria macrospora were identical to those of strain CBS 346.62 (MH858175 in ITS, MH869769 in 28S rDNA). The sequence of β-tubulin of W471 was identical to that of strain FGSC 4818 (FR774340) belonging to species S. macrospora. With β-tubulin sequences, S. macrospora can be distinguished from the other known Sordaria species, whereas its ITS and 28S rDNA sequences were identical to those of S. lappae and S. fimicola. The isolate W913 of the species Mortierella biramosa matched the CBS 370.95 strain (JX976094 in ITS, HQ667389 in 28S) with sequence similarities of 100% (555/555 bp) for ITS and 99.9% (872/873 bp) for 28S rDNA sequences.
Phylogenetic relationships between the Korean and other previously published authentic isolates were analyzed using ME and ML analyses of the ITS rDNA sequences for Paraphoma (Fig. 1A) and Mortierella (Fig. 1B), and the concatenated alignment of ITS and 28S rDNA sequences for Sordaria (Fig. 1C). In Fig. 1, only the ME tree is shown because the topologies constructed from both analyses are congruent. The Korean isolate W251 was grouped with previously published isolates of species Paraphoma radicina, including the epitype strain of CBS 11179, with high bootstrap values of 100% for ME and 99% for ML. In the BLASTn search results, the ITS sequence of the Korean isolate W251 was identical to that of strain CBS 111.79 (NR_156556). The ITS sequence of isolate W913 was identical to that of the authentic strain CBS 370.95 (JX976094) of species Mortierella biramosa, and the two strains were grouped together in the phylogenetic tree. This grouping was supported by high bootstrap values of 100% for ME and 99% for ML. The Korean isolate W471 was grouped with three Sordaria species, namely S. macrospora, S. lappae, and S. fimicola in the tree inferred from a concatenated alignment of ITS and 28S rDNA sequences. In the β-tubulin tree (Fig. 1D) for the indistinguishable taxa in the ITS and 28S tree (in the blue box in Fig. 1C), the isolate W471 was grouped with S. macrospora with a high bootstrap value of 99% in ME and ML, and was distinguishable from S. lappae and S. fimicola.

Fig. 1. Minimum evolution phylogenetic trees of (A) Paraphoma, (B) Mortierella species based on ITS rDNA sequences, (C) Sordaria species based on a combined dataset of internal transcribed spacer (ITS) and 28S rDNA sequences, and (D) β-tubulin sequences of Sordaria species in a blue box of (C). Bootstrapping values of minimum evolution and maximum likelihood, higher than 70%, are presented above the branches (1,000 replicates). The scale bar represents the number of nucleotide substitutions per site. The strains that were collected in Korea are shown in bold.
Taxonomy
Paraphoma radicina (McAlpine) Morgan-Jones & J.F. White (1983) [MB#109141] (Fig. 2A-J)
Description: Colonies formed a radiate pattern with short and dense aerial mycelia on PDA, V8A, and CMA at 25℃. Aerial mycelium white to pale grey. Pycnidia setose; globose to subglobose; brown to blackish-brown; non-papillate or papillate ostioles; 180-450 µm in diameter. Conidiogenous cells bottle-shaped; 5-7×3-7 µm. Conidia aseptate; ellipsoidal to subglobose; 3-6×1-3 µm.
Isolate examined: Korea, Gwangju; Gwangsan-gu; Donggok-dong (35°04'59.4"N 126°46'35.3"E), ex freshwater, June 29, 2016, Y.-J. Choi (NNIBRFG31692, W251).
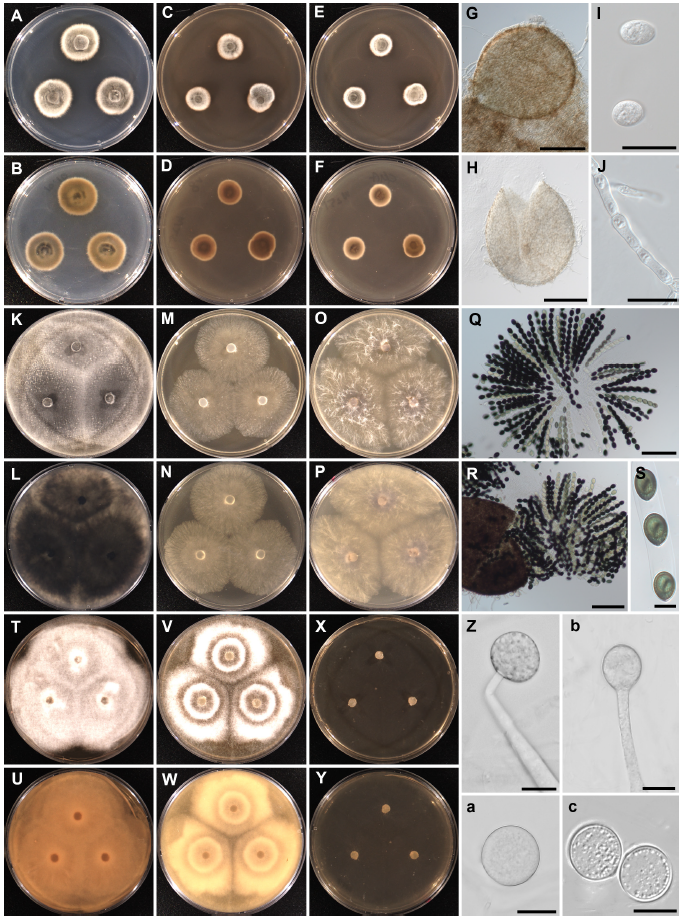
Fig. 2.Cultural and morphological characteristics of Paraphoma radicina NNIBRFG31692 (W251; A-J), Sordaria macrospora NNIBRFG31693 (W471; K-S), and Mortierella biramosa NNIBRFG24241 (W913; T-c). Colonies on potato dextrose agar (first column), V8 juice agar (second column), corn meal agar (third column) after incubation for 72 h at 25℃; upper at obverse view and lower at reverse view. Paraphoma radicina: pycnidia (G, H), conidia (I), and conidiogenous cells (J) (scale bars: G, H=100 μm, I, J=10 μm). Sordaria macrospora: rosettes of asci (Q), opening of perithecium (R) and ascospores (S) (scale bars: Q, R=100 μm, S=20 μm). Mortierella biramosa: sporangium on the sporangiophores (Z, b, a) and chlamydospores (c). Scale bars : 20 μm.
Sordaria macrospora Auersw. (1866) [MB#237763] (Fig. 2K-S)
Description: Sparse aerial mycelia of pale white color formed on the PDA, V8A, and CMA. After two weeks, dark pycnidia and perithecia formed on the PDA. Vegetative hyphae hyaline; up to 5 μm wide. Perithecia pyriform; setose; 370-400×250-300 µm. Setae were scarce; smooth-walled. Asci with truncate apex and apical rings unitunicate; aseptate, cylindrical; 8 spored, 160-175×ca. 20 µm in size; formed rosettes. Ascospores uniseriate; linearly arranged; green to pale brown, later brown; 22-35×15-20 µm.
Isolate examined: Korea, Jeollabuk-do; Gimje-si; Mangyeong-eup (35°51'10.0"N 126°49'22.2"E), ex clamshell, Jan. 4, 2017, Y.-J. Choi (NNIBRFG31693, W471).
Mortierella biramosa Tiegh. (1875) [MB#240881] (Fig. 2T-c)
Description: Colonies formed a radiate pattern with cottony aerial mycelia on PDA and V8A. On CMA showed submerged growth. Colony diameter on PDA, V8A, and CMA >70 mm at 25℃, after 72 h. Sporangiophores 300-1,500 µm long. Sporangia globose; 30-40 µm in diameter. Sporangiospores globose; smooth-walled with granular contents 5-8 µm in diameter. Chlamydospores globose; 15-35 µm in diameter.
Isolate examined: Korea, Jeollanam-do, Boseong-gun, Ungchi-myeon (34°41'51.3"N 126°59'21.9"E), ex soil sediment, May 15, 2019, B. Nam and Y.-J. Choi (NNIBRFG24241, W913).
Discussion
Cultural, morphological, and phylogenetic characteristics of the isolate W251 was identical to Paraphoma radicina (McAlpine) Morgan-Jones & J.F. White [24], which is the type species for the genus Paraphoma. This genus was initially introduced as a section of Phoma but later resurrected as a distinct genus. It is differentiated by its setose pycnidia and dictyochlamydospores [24,25]. Paraphoma have been predominantly isolated from the rhizosphere and the phyllosphere of crops and trees, with several species being identified as plant pathogens [25-27], but also reported in freshwater [28-30] and brackish [31] habitats. P. radicina has been isolated from wood and herbaceous debris submerged in aquatic ecosystems [29-31]. The Korean isolate W251 was isolated from reservoir water. P. radicina produces chemical compounds for antimicrobial activity against bacteria and fungi [28,29]. El-Elimat et al. [29] identified nine compounds belonging to isochromenones, isobenzofuranones, and tetrahydronaphthalenes from P. radicina isolated from wood in a lake, and then evaluated their antimicrobial activity. Of these compounds, clearanol C and isobenzofuranone showed promising antimicrobial activity against Staphylococcus aureus and Candida albicans, respectively. P. radicina could be useful as a bioresource in bioindustry. The presence of P. radicina in freshwater habitat is also remarkable. This species may play an ecologically important role in biogeochemical cycles in freshwater ecosystems, as they are distributed across a diverse range of sources, such as plant debris and water.
Isolate W471 obtained from shellfish on the reservoir shore was consistent with the cultural, morphological, and phylogenetic descriptions of Sordaria macrospora Auersw. [32,33]. This species serves as a model organism for studying fungal sexual fruiting body formation [34]. The genus Sordaria has been reported in freshwater environments [35,36] as well as in wood [37-39] and grass [40], and other terrestrial plants. S. macrospora is a well-known coprophilous fungus that is commonly isolated from herbivore dung [41]. However, the Korean isolate W471 was found in clam shells in the reservoir riparian zone. To our knowledge, S. macrospora inhabiting freshwater mollusks has not been previously recorded. Previous studies have reported shellfish-derived fungi focusing on pathogenic fungi, such as Aspergillus, Fusarium, and Penicillium, isolated from marine bivalve mollusks [42-44], whereas saprophytic fungi associated with freshwater bivalves have been poorly studied. There is a lack of research on the role of fungi in the metabolism of bivalves, although the presence of fungi in mollusks has long been known [45]. Future studies are warranted to investigate the ecological role of fungi inhabiting bivalve mollusks. The class Sordariomycetes, including the genus Sordaria, is a key freshwater fungal group, which accounts for half of the total known fungi in freshwater ecosystems [35,46]. This study demonstrated the presence of Sordaria in clam shells. Their ecological functions should be studied further to understand the relationship between Sordaria and shellfish.
Isolate W913 was identified as Mortierella biramosa Tiegh. based on their cultural, morphological, and molecular phylogenetic analyses [47-49]. This strain was found from the soil sediment in the reservoir in the present study. The genus Mortierella is commonly found in terrestrial ecosystems and is known for being beneficial in promoting plant growth [50-52]. Some members of Mortierella produce an important polyunsaturated fatty acid and arachidonic acid (ω-6,5,8,11,14-cis-eicosatetraenoic acid; ARA). They have diverse functions in the human body and broad applications in industrial fields [53]. This group is also distributed in freshwater ecosystems, including water, plants, and soil [54,55], with a remarkable capacity to decompose organic matter [56]. Mortierella elongata, M. horticola, M. humilis [55], and a new species, M. fluviae [57], were reported in a freshwater sample in Korea. M. biramosa W913 obtained in the present study was also isolated from the littoral zones of the reservoir. The presence of Mortierella in freshwater has often been documented, but their ecological roles are still unknown. The Mortierella species in freshwater ecosystems may be highly valuable. Therefore, further research is required to analyze their biological activities.
Given the biochemical and biophysical distinctions highlighted in previous studies, these isolates could be used as bioresources. Therefore, this study provides essential information for future studies on the potential use of these isolates as biological materials.




