INTRODUCTION
Gotjawal, which is a compound word comprising “Got” (meaning forest) and “jawal” (meaning a land scattered with stones and pebbles in Jeju), is the name of a forest in South Korea with irregularly scattered volcanic rock masses [1]. Gotjawal is divided into four terrains: Hangyeong-Andeok, Aewol, JocheonHamdeok, and Gujwa-Seongsan Gotjawal [2]. Moreover, Gotjawal is recognized worldwide for its high ecological value because this region has diverse ecosystems that host complex biodiversity [3,4].
Although the diversity of mushroom species is expected to be high in Gotjawal, research on unrecorded species has mainly been conducted only in the Jocheon-Hamdeok terrain; thus, further research in other Gotjawal terrains is required in this aspect [5-9].
Here, we describe one specimen each from the genera Gymnopilus P. Karst. (1879; Hymenogastraceae), Marasmius Fr. (1836; Marasmiaceae), and Mycena (Pers.) Roussel (1806; Mycenaceae), which has not been previously recorded in Hwasun Gotjawal and Jeju Gotjawal Provincial Park of the HangyeongAndeok terrain, by comparing the morphological characteristics and molecular analysis results of species. The results serve as baseline data that can be used to describe the diversity of mushroom species in Gotjawal.
MATERIALS AND METHODS
Taxon sampling
The specimens used in this study were collected from Hawsun Gotjawal, Seogwipo, Jeju, South Korea, in 2020. A D700 camera (Nikon, Tokyo, Japan) was used to capture images of fruit bodies. The size of the fruit bodies was measured indoors after recording the location information and field substrate. The specimens were deposited in the herbarium of the Biodiversity Research Institute, Jeju Technopark, South Korea, where a constant temperature (25℃) and humidity (50%) were maintained after drying at 60℃ for 24 hr.
Macroscopic and microscopic characteristics
Macroscopic characteristics, such as the size, shape, and color of the pileus, lamellae, and stipe, were observed and described based on captured images and dried specimens. The color was analyzed using the Korea Standard Color Analysis (KSCA) and was indicated with the KS symbol [10]. Microscopic features, including the size, shape, and thickness of basidia, basidiospores, cheilocystidia, pleurocystidia, as well as the number of sterigmata of basidia, were observed and described using an Eclipse 80i microscope (Nikon, Tokyo, Japan). The samples were stained with 1% Congo red and 1% phloxine and then washed with 3% potassium hydroxide.
DNA extraction and sequencing
Genomic DNA was extracted from dried specimens weighing approximately 100 mg using a 5-min Mushroom DNA Extraction Kit (Catalogue number D-1016, Biofactories, Los Angeles, CA, USA), following the manufacturer’s protocol. The universal primer sets ITS1 and ITS4 [11] were used to amplify the internal transcribed spacer (ITS) region. Subsequently, polymerase chain reaction and sequencing of the amplified products were performed by Bionics Inc. (Seoul, South Korea). The resulting sequences were compared with the ITS sequences in GenBank using the Basic Local Alignment Search Tool provided by the National Center for Biotechnology Information.
Molecular phylogenetic analyses
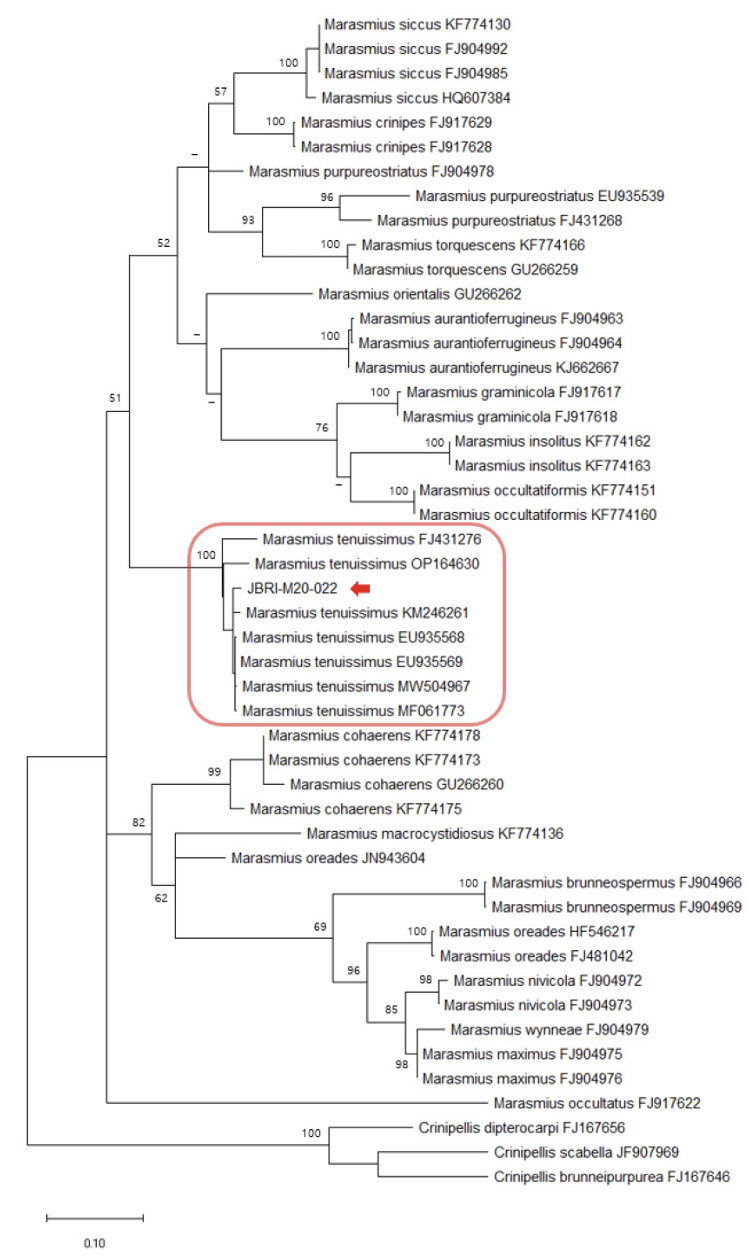
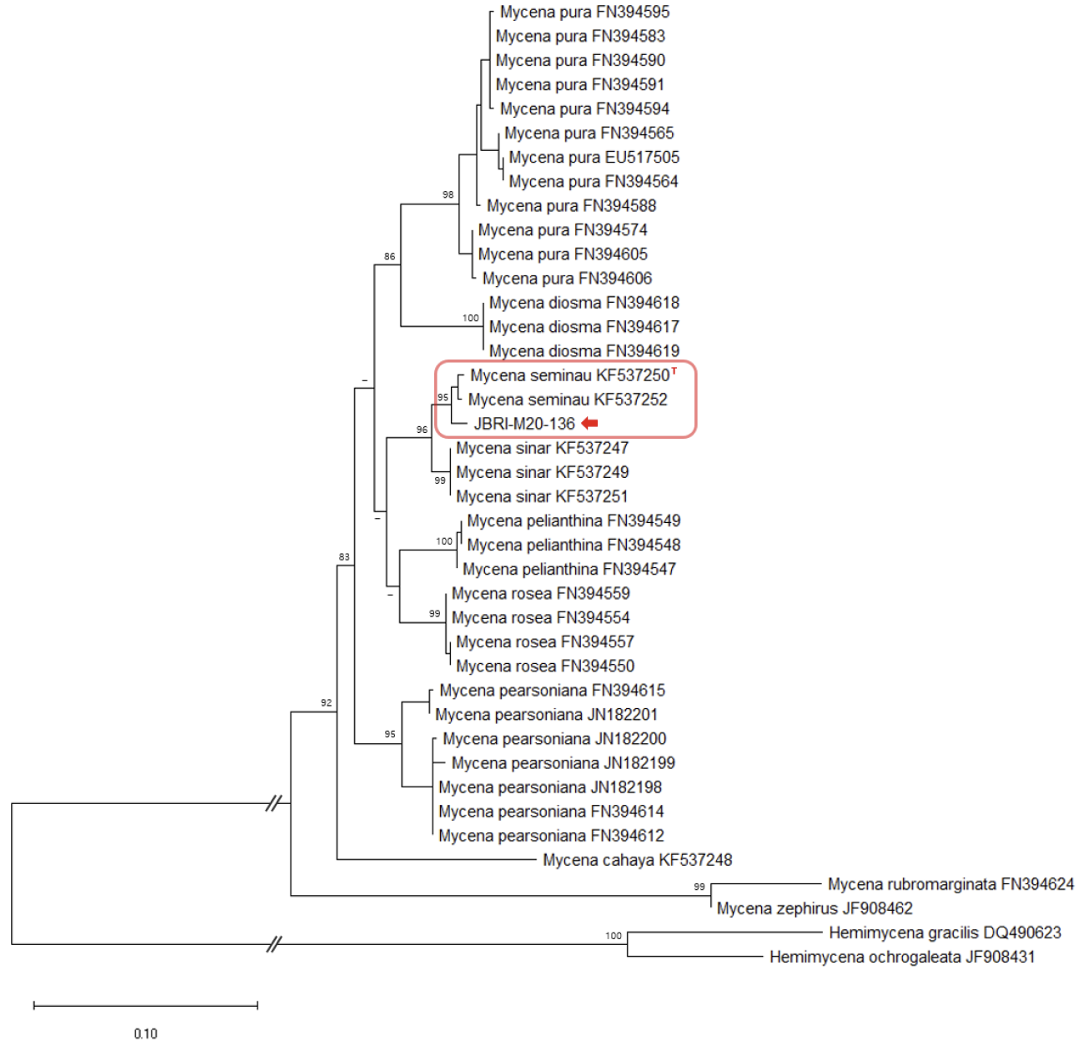
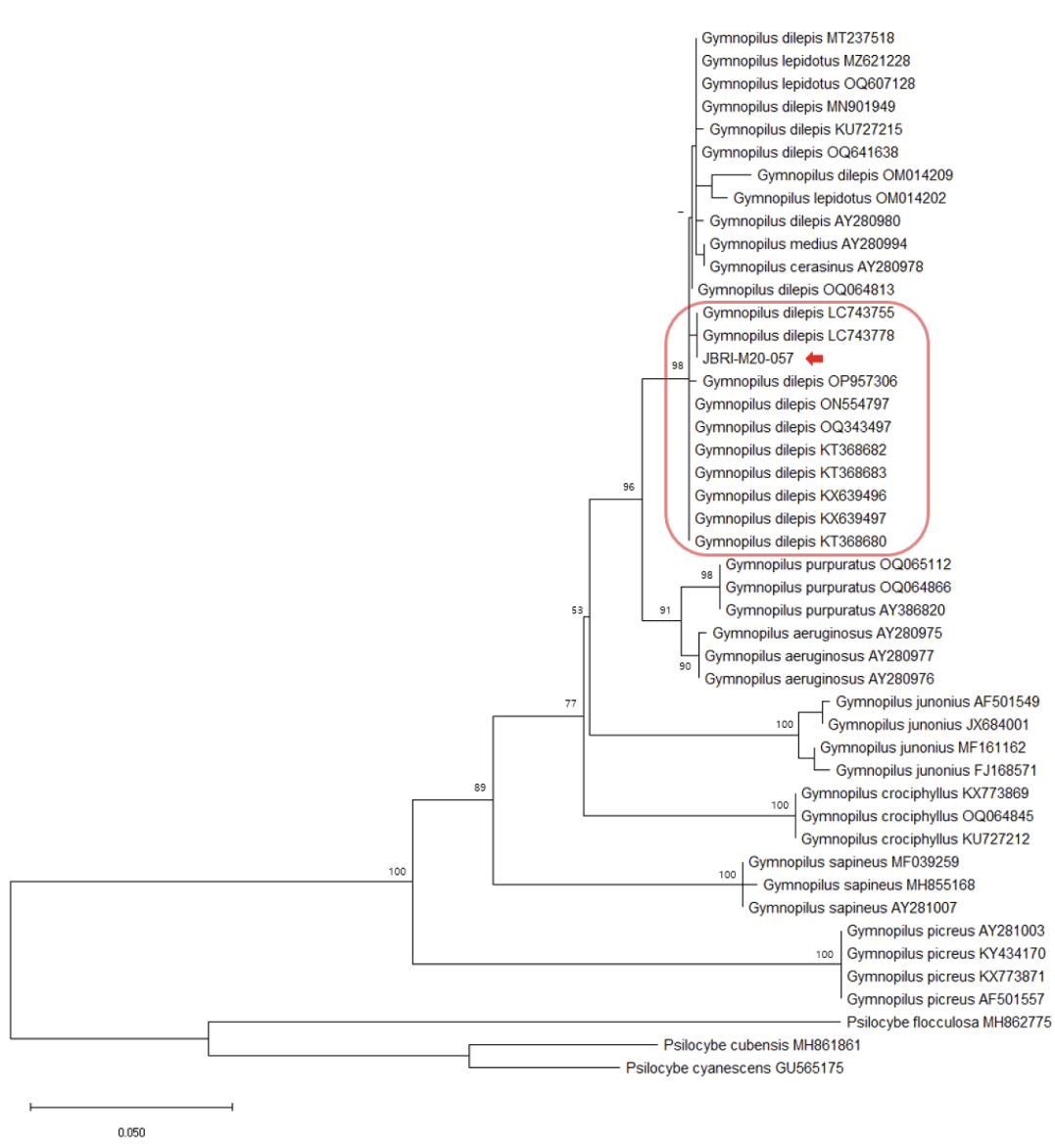
The ITS sequences from GenBank were used to analyze phylogenetic relationships, referencing Kiyashko et al. [12] and Chew et al. [13]. Sequences were aligned using the MEGA X software [14]. A maximum likelihood (ML) tree was constructed using the general time-reversible (GTR) model combined with the gamma distribution (G) and invariant sites (I) model. The GTR+G+I model was selected as the best evolutionary model for ML by jmodeltest2 [15]. Bootstrap testing was performed with 1,000 replicates. The accession numbers of the GenBank sequences, along with their respective scientific names, are provided in Figs 1, 2, and 3.
Fig. 1
Maximum likelihood (ML) tree of internal transcribed spacer (ITS) sequences of Gymnopilus. The bootstrap values above 50% are shown next to the branches (1,000 replicates). JBRI-M20-057 is the accession number of our specimen.
RESULTS
Comparison of the ITS sequences from our collected Gymnopilus, Marasmius, and Mycena specimens with other sequences from GenBank revealed 100.0%, 99.1%, and 98.9% homology to G. dilepis (LC743755), Ma. tenuissimus (EU935568), and My. seminau (KF537250), respectively.
The phylogenetic analysis further confirmed the grouping of the Gymnopilus, Marasmius, and Mycena species with G. dilepis, Ma. tenuissimus, and My. seminau, respectively (Figs 1, 2, and 3).
Taxonomy
Gymnopilus dilepis (Berk. & Broome) Singer, Lilloa 22:560, 1949 [16] (Fig. 4)
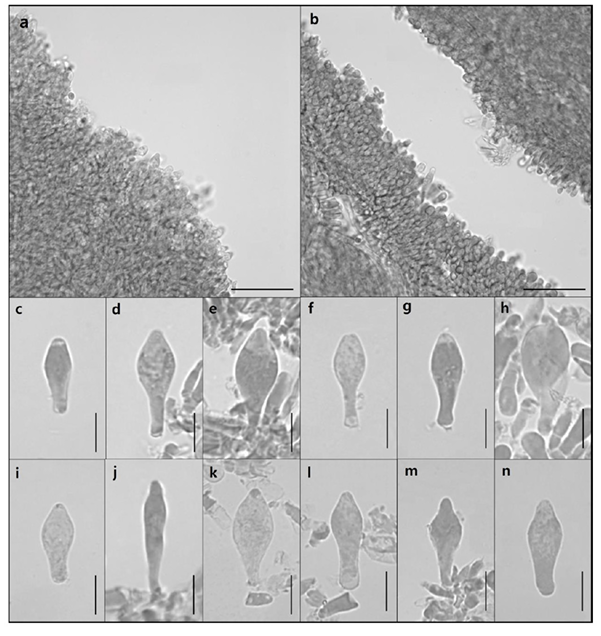
Pileus (Fig. 4a) 12.10–24.90 mm in diameter, convex shape, surface squamulose, orange (KS 1.25YR 7/10) to dark red (KS 8.75R 4/10) toward the center; Lamellae (Fig. 4b) lamellulae in between, pale orange (KS 5YR 8/4); Stipe (Fig. 4) 33.68–38.71 mm, central, cylindrical, pinstripe, pale orange (KS 5YR 8/4) to pale red (KS 10R 7/6); Basidia (Fig. 5a–5j) 6.81–9.07 × 19.85–33.99 µm, clavate with thin walls, hyaline, four sterigmata with spores at the tip; Basidiospores (Fig. 5k and 5l) 5.36–6.94 × 3.78–4.70 µm, elliptical with thick walls, verrucose; Cheilocystidia (Fig. 6a, 6c–6h) 6.88–12.27 × 19.39–28.83 µm, clavate with a thin handle, thin-walled; Pleurocystidia (Fig. 6b, 6i–6n) 4.99–9.98 × 20.08–29.29 µm, clavate with a thin handle, thin-walled.
Habitat: Occurred on dead wood. Hwasun Gotjawal, the collection site of D. dilepis was covered by both evergreen and deciduous broad-leaved trees.
Specimen examined: South Korea, Jeju, Seogwipo, Hwasun Gotjawal (33°15'55.8792" N, 126° 19'51.9564" E), June 19, 2020, S. H. Lee (JBRI-M20-057; GenBank accession number OR177789).
Remarks: Contrary to the pleurocystidia observed in this study, they were described as absent by Suwannarach et al. [17]. Additionally, Thomas et al. [18] reported that pleurocystidia are absent or extremely rare.
Marasmius tenuissimus (Sacc.) Singer, Fl. Neotrop. Monogr. 17: 258, 1976 [19] (Fig. 7)
Pileus (Fig. 7a) up to 7.56 × 4.57 mm, convex to plano-convex, sulcate, dull, dry, glabrous, pale yellow (KS 10Y 8/4) to brown (KS 1.25Y 6/8); Lamellae (Fig. 7b) adnate, distant, intervenose, white yellowish green (KS 5GY 9/2); Stipe (Fig. 7b) 0.69 × 1.71 mm, lateral, cylindrical, velutinous, dark brown (KS
Fig. 4
Fruit body images of the G. dilepis specimen in the field, Hwasun Gotjawal. a, b: JBRI-M20-057 (Scale bars = 10 mm).

Fig. 5
Microscopic images of basidia and basidiospores of G. dilepis. a–j: Basidia with sterigmata. k, l: Basidiospores (Scale bar = 10 µm).
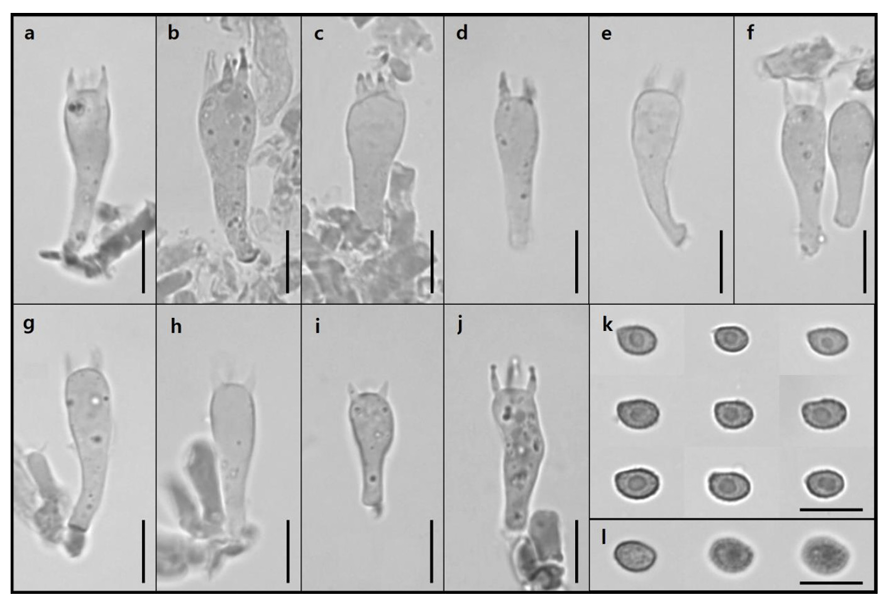
Fig. 6
Microscopic images of cheilocystidia and pleurocystidia of G. dilepis. a, c–h: Cheilocystidia. b, i–n: Pleurocystidia (Scale bar: a, b = 50 µm; c–n = 10 µm).
5Y 2/2); Basidia (Fig. 8c–8g) 7.74–9.48 × 32.40–43.07 µm, clavate with thin walls, 4 sterigmata with spores at the tip; Basidiospores (Fig. 8b) 11.42–13.28 × 6.06–6.77 µm, elliptical with thin walls, smooth; Cheilocystidia (Fig. 9) 7.18–21.72 × 19.17–44.06 µm, Siccus-type broom cells, cylindrical to clavate, hyaline, thin-walled; Pleurocystidia absent.
Habitat: Scattered or gregarious on branches. Jeju Gotjawal Provincial Park, the collection site for Ma. tenuissimus is covered by both evergreen and deciduous broad-leaved trees.
Specimen examined: South Korea, Jeju, Seogwipo, Jeju Gotjawal Provincial Park (33°17'3.3108" N, 126°16'19.7616" E), May 19, 2020, S. H. Lee (JBRI-M20-022; GenBank accession number OR177790).
Remarks: The specimen in this study was not sufficiently mature; therefore, the size of the pileus was smaller than that reported in other studies [19-21], and it was difficult to observe basidia with sterigmata.
Mycena seminau A.L.C. Chew & Desjardin Mycologia 106(5): 985, 2014 [13] (Figure 10)
Pileus (Fig. 10a and 10c) 11.13–25.42 mm in diameter, convex to flat shape, pressed apex, smooth and crumpled surface without gloss, light brown (KS 5YR 8/6) to dark brown (KS 8.75YR 2/2), sometimes darker toward the edge. Lamellae (Fig. 10b) lamellulae in between, grayish brown (KS 5YR 5/2). Stipe (Fig. 10c) 36.81–62.74 mm, central, cylindrical, smooth and pruinose, pinstriped, basal tomentum, yellowish-brown (KS 8.75YR 7/4) to dark brown (KS 1.25YR 3/6), and darker toward the top. Basidia (Fig. 11d–11m) 6.59–8.75 × 29.63–42.97 µm, clavate with thin walls, 2 or 4 sterigmata with spores at the tip. Basidiospores (Fig. 11c) 3.92–5.52 × 7.36–10.61 µm, elliptical with thin walls, smooth, small apiculus. Cheilocystidia (Fig. 12a and 12d–12h) 5.17–12.61 × 19.76–51.02 µm, multiform, elliptical to clavate or lageniform, sometimes mucronate, thin-walled. Pleurocystidia (Fig. 12b and 12c) 9.05–9.47 × 19.11–23.62 µm, very rare or absent, only elliptical and lageniform observed, thin-walled.
Habitat: Scattered or gregarious on humus. Hwasun Gotjawal, the collection site for My. seminau was covered by both evergreen and deciduous broad-leaved trees.
Specimen examined: South Korea, Jeju, Seogwipo, Hwasun Gotjawal (33°15'59.3568" N, 126° 19'56.0748" E), September 22, 2020, S. H. Lee (JBRI-M20-136; GenBank accession number OM109694).
Remarks: “Seminau” means shine in Malay. It refers to the bioluminescence of this species [13].
Fig. 7
Fruit body images of the Ma. tenuissimus specimen in the field, Jeju Gotjawal Provincial Park. a, b: JBRI-M20-022 (Scale bars = 10 mm).

Fig. 8
Microscopic images of basidia and basidiospores of Ma. tenuissimus. a: Clustered basidia. b: Basidiospore. c–g: Basidia with sterigmata (Scale bar = 10 µm).
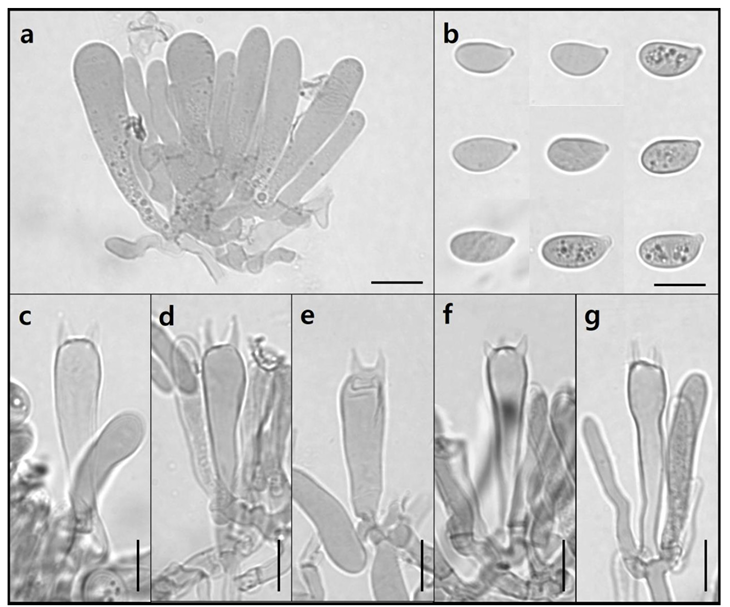
Fig. 9
Microscopic images of cheilocystidia of Ma. tenuissimus. Pleurocystidia are absent (Scale bar = 10 µm).
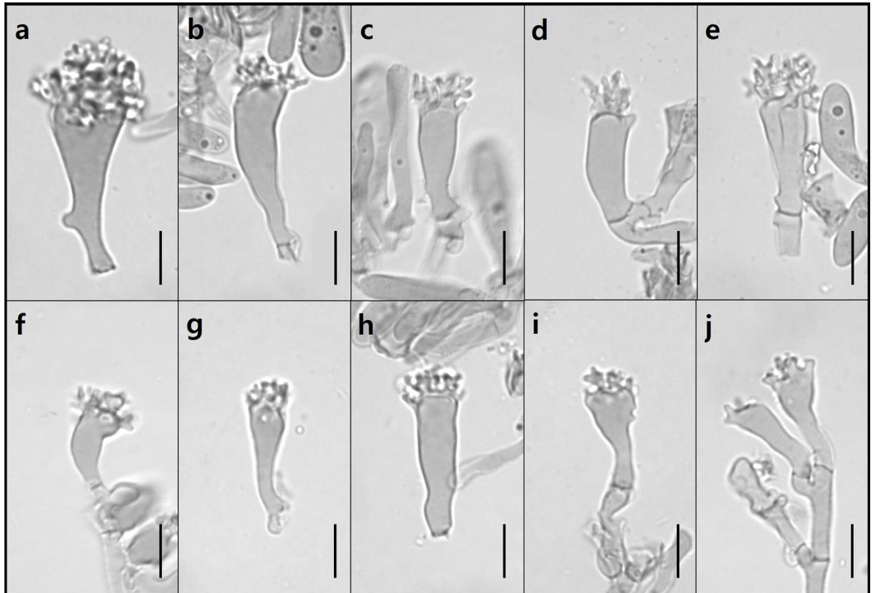
Fig. 10
Fruit body images of the My. seminau specimen in the field, Hwasun Gotjawal. a–c: JBRI-M20-136 (Scale bars = 10 mm).10 µm.

DISCUSSION
In this study, morphological and molecular phylogenetic analyses identified one specimen each of the genera Gymnopilus, Marasmius, and Mycenaas an unrecorded species in South Korea.
Morphological analysis of the Gymnopilus specimen showed microstructural characteristics similar to those of G. dilepis [17,18,22,23]. Contrary to the description by Thomas et al. [18], who stated that pleurocystidia is absent or extremely rare, several pleurocystidia were observed in this study [22]. In addition, G. dilepis was described as having a sulfur-colored pileus in the original study and an orangecolored pileus in other studies; however, the specimen in this study had a reddish pileus [17,18,24]. The Marasmius specimen examined in this study was immature; therefore, the size of the pileus was smaller than that in other studies, but the microstructure size was generally larger [19-21,25]. The microstructure size in the study by Wannathes et al. [21] was the most similar to that of the specimen in this study; however, the cheilocystidia shape differed slightly.In the Mycena specimen, the appearance and microstructure were similar to those in the original description by Chew et al. [13]. One difference was that the pleurocystidia were slightly smaller than those in the original description. This may be due to insufficient data because of the difficulty in observing pleurocystidia. Additionally, the basidia observed in our specimens were larger than those observed in the original description.
Molecular analyses revealed that the ITS sequence of the Gymnopilus specimen showed 100.0% homology with G. dilepis (LC743755) and was included in the G. dilepis group of the phylogenetic tree with a high bootstrap value (Fig. 1). In Figure 1, our specimen is grouped into one clade with G. dilepis, G. lepidotus, G. medius, and G. cerasinus. According to Laura et al. [26], these four species belong to the lepidotus-subearlei clade, and species within this clade have erect, reddish squamules on the pileus. However, as G. cerasinus has recently been synonymized with Pholiota discolor, and Thorn et al. [27] mentioned that the sequence of G. cerasinus (AY280978) matches that of G. lepidotus, it seems that AY280978 might have incorrectly registered the sequence of G. lepidotus as G. cerasinus. Additionally, Thomas et al. [18] noted that G. lepidotus is morphologically very similar to G. dilepis and might be synonymized in the future. Therefore, further research into the morphological and genetic characteristics of these four species seems necessary.The ITS sequences of the Marasmius and Mycena specimens showed 99.1% homology with Ma. tenuissimus (EU935568) and 98.9% homology with My. seminau (KF537250), respectively. They were also included in the Ma. tenuissimus and My. seminau groups of each phylogenetic tree with high bootstrap values (Figs. 2 and 3).
Most morphological characteristics of Gymnopilus, Marasmius, and Mycena specimens were similar to those described in other studies, with only slight differences, and the molecular analysis results supported the morphological identification.
Therefore, herein, we report three species, G. dilepis, Ma. tenuissimus, and My. seminau, which have not been recorded previously in South Korea.
Additionally, the number of mushroom species reported in Jeju Island stands at 82 families, 293 genera, 841 species, as announced by the Jeju Special Self-Governing Province in 2019 [28]. Since that announcement, new and previously unrecorded species have been discovered in Jeju. With the addition of regional survey data, including the three unrecorded species mentioned in this study, there appears to be a need to update the tally of mushroom species on Jeju Island.