INTRODUCTION
Meju is a fermented soybean block used as a key material for preparing soy sauce (ganjang) and soybean paste (doenjang), the popular Korean traditionally fermented foods [1]. Meju is produced from soybeans rich in nutrients, including proteins, carbohydrates, and vitamins; hence it has been a good source of proteins and fat for a long time. Furthermore, it contains numerous essential amino acids and functional materials that are essential for physiological regulation of the body and prevent various diseases such as cardiovascular disorders, cancer, and atopic dermatitis [2].
Meju produced using the traditional procedure is generally prepared by washing soybeans, steeping, steaming, cooling, mashing, molding, drying, and fermentation. During the natural drying and fermentation process of meju, various microorganisms such as bacteria, yeasts, and mold, present in soil or rice straw, are involved in fermentation. These microorganisms can produce a variety of compounds that affect the unique taste and flavor of meju [3]. Therefore, the microbial community is important for the quality of meju and other fermented foods. Traditionally produced meju has the advantage of enabling the production of unique aroma and taste; however, its disadvantage is that the quality is not constant and depends on the fermentation conditions, including temperature, humidity, and time [4]. When meju is contaminated with foodborne bacteria or toxin-producing fungi during fermentation, it can cause various diseases, including food poisoning and cancer. Therefore, it is important to control the microbial community during meju fermentation.
Fungi play a key role in the fermentation of meju [5,6]. Fungi produce and secrete various hydrolytic enzymes such as amylase, protease, and lipase, decomposing macromolecules such as starch and proteins of soybeans to produce the unique taste and flavor [7]. According to previous studies, the major fungal genera involved in the fermentation process include Aspergillus, Rhizopus, Mucor, and Penicillium [6-9]. In particular, Aspergillus species belonging to Aspergillus section Flavi, including A. oryzae and A. sojae, are predominant and key microorganisms for meju fermentation [6,9]. These fungi are nontoxigenic and detoxify during the fermentation process. A. oryzae is dominant during the fermentation of meju, producing reducing sugars, amino acids, and metabolites to improve aroma and taste [6]. A. oryzae is nontoxigenic and cannot produce the carcinogenic mycotoxin aflatoxin B1 [10,11], which is because it lacks the aflatoxin gene cluster. Moreover, A. oryzae detoxifies various mycotoxins, including aflatoxins and ochratoxins [12,13]. Therefore, A. oryzae, also known as yellow koji mold, has been used for meju fermentation and might act as a starter for fermentation.
In this study, we examined the distribution of fungal communities in meju samples during fermentation and final production using metagenomic and morphological analyses. Among the fungal isolates, we investigated the ability to produce amylase, protease, and aflatoxin and propose several candidates that can be used as starters for meju fermentation.
MATERIALS AND METHODS
Isolation and identification of fungal isolates from meju
Meju samples were collected from Andong Jebiwon Traditional Food Co., Ltd. (Andong, Korea). Meju bricks were collected in the middle stage of meju fermentation (Meju M) or the final fermented meju products (Meju F). After freeze-drying the meju bricks, samples (5 g) were added to sterilized water (45 mL), homogenized, and then serially diluted using sterilized water. The diluted samples were inoculated onto potato dextrose agar (PDA, BD, NJ, USA) containing 100 μg/mL chloramphenicol (Sigma, MO, USA) and cultured for 3 days at 30℃. To further identify the fungal isolates, 100 isolates were randomly selected from each sample and inoculated into PDA, and then the agar plate was cultured for 5 days at 30℃. The obtained isolates were grouped according to their morphological characteristics, and three representative strains from each group were used for additional experiments. To identify the representative isolates, the genomic DNA of each strain was extracted as described previously [14]. The internal transcribed spacer (ITS) region of each sample was amplified using ITS1 (5ʹ-GTCAAACCCGGTCATTTAGAGG-3ʹ) and ITS4 (5ʹ-CTTCACTCGCCGTTACTGAG-3ʹ) primers and 2× Go tag master mix (Promega, WI, USA). The polymerase chain reaction (PCR) products were sequenced at SolGent Co., Ltd. (Daejeon, Korea), and the sequence data were further analyzed using the NCBI BLAST server (National Center for Biotechnology Information, Meryland, Bethesda, USA). A phylogenic tree was generated using the MEGA X program (https://www.megasoftware.net/).
Metagenomic analysis
Total genomic DNA was extracted from each freeze-dried meju sample using the Powersoil kit (QIAGEN, Hilden, Germany). Library was prepared using the Nextra XT index kit (Illumina, CA, USA) and sequenced on an Illumina MiSeq instrument. The sequencing results were further processed using CutAdapt v1.11 and assembled into one sequence using FLASH v1.2.11. The processed data were clustered through Cd-hit v4.6 (identify >99%, coverage >80%), and taxonomy profiling was performed using NCBI Megablast to obtain a heatmap for the distribution of fungi in meju [12,15,16]. Sequencing and data analysis were performed at Teragen Bio Inc. (Seongnam, Korea).
Aflatoxin B1 analysis
The production of aflatoxin B1 was evaluated using thin-layer chromatography (TLC) as described previously [15]. Each strain was inoculated into complete medium (CM, 20 g glucose, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20× nitrate salt solution, 1 mL of 1,000× trace element per 1 L) and cultured for 7 days at 30℃ under dark conditions. Next, 5 mL of chloroform (Duksan, Ansan, Korea) was added to the culture media and mixed vigorously. The organic solvent layer was separated by centrifugation and filtered using Whatman No. 4 (Sigma). The dried sample was resuspended with 100 µL of chloroform and spotted onto a TLC silica plate (Kiesel gel 60, 0.25 mm; Merck). The plate was placed in a chamber saturated with a solvent of chloroform:acetone at 9:1 ratio (Duksan) and exposed to ultraviolet (UV) light (366 and 254 nm).
Analysis of the aflatoxin biosynthesis gene cluster
PCR and gel electrophoresis analyses were conducted to check the aflatoxin biosynthesis gene cluster and the aflatoxin-producing ability of A. flavus/oryzae strains. For the control, the aflY region containing both aflatoxin-producing and nonproducing strains was amplified by PCR using the primers aflY_F (5ʹ-CATGTGCATCACGGCATGAG-3ʹ) and aflY_R (5ʹ-ATAACCAGCGGTTCTGCCAA3ʹ). The aflF-aflU interregion, the breaking point pattern, was amplified by PCR using the primers aflF_F (5ʹ-CAACTGCTCGACTGTCGTCT-3ʹ) and aflU_R (5ʹ-GAATTGGCTTCGCTCCCTCT-3ʹ). The size of the PCR product was confirmed by electrophoresis.
Analyses of amylase and protease activities
The amylase activity was evaluated as described previously [16]. Each strain was inoculated into CM containing modified starch (20 g starch, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20 × nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Next, 2 mL Lugol solution (Sigma) was added to the plate to induce a blue–violet iodinestarch reaction. After removing the Logol solution, the width of the clear zone at the end of the mycelium was measured. For analyzing the activity of protease, each strain was inoculated into CM containing modified skim milk (20 g skim milk, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20× nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Then, the clear zone was measured.
RESULTS
Evaluation of fungal distribution in meju using metagenomic analysis
Meju brick samples were collected from the middle stage of meju fermentation (Meju M) or the final fermented meju products (Meju F). DNA samples were sequenced and analyzed for fungal population. Analysis of the fungal community at the genus level revealed the distribution of Aspergillus, Pichia, Rhizopus, and Lichtheimia. Aspergillus spp. were the most dominant in all meju samples. Pichia spp. were found in Meju M samples (8.3%), but their composition decreased in Meju F samples (0.06%). Rhizopus and Lichtheimia spp. were not detected in Meju M samples, but their distribution was increased in Meju F samples (Fig. 1).
Identification of fungi in meju using culture-based methods
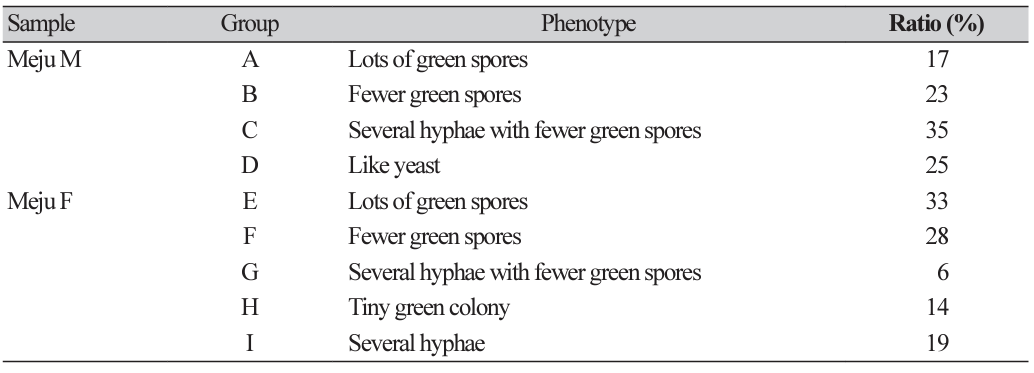
Samples collected from Meju M and Meju F were suspended using water, diluted, and spread onto PDA plates containing chloramphenicol. The plates were incubated for 5 days, after which 100 isolates were randomly selected, inoculated into new PDA media, and cultured for 3 days at 30℃. Colonies with similar morphological phenotypes were grouped from both Meju M (Groups A-D) and Meju F (Groups E-I) samples (Fig. 2A). Among Meju M samples, fungi belonging to Group A produce numerous green spores, fungi belonging to Group B produce fewer green spores, fungi belonging to Group C produce numerous hyphae with fewer green spores, and fungi belonging to Group D appears as a yeast colony. Among Meju F samples, fungi belonging to Group E produce several green spores, fungi belonging to Group F produce fewer green spores, fungi belonging to Group G produce several hyphae with fewer green spores, fungi belonging to Group H appears as a tiny green colony, and fungi belonging to Group I produce several hyphae (Fig. 2A).
Fig. 1
Metagenomic analysis of fungal community distribution in the meju samples. (A) Images of meju bricks. Meju M (sample of the middle stage of meju fermentation) and Meju F (the final fermented meju products). (B) The heatmap shows the relative comparison of the distribution of fungal communities between Meju M and Meju F.
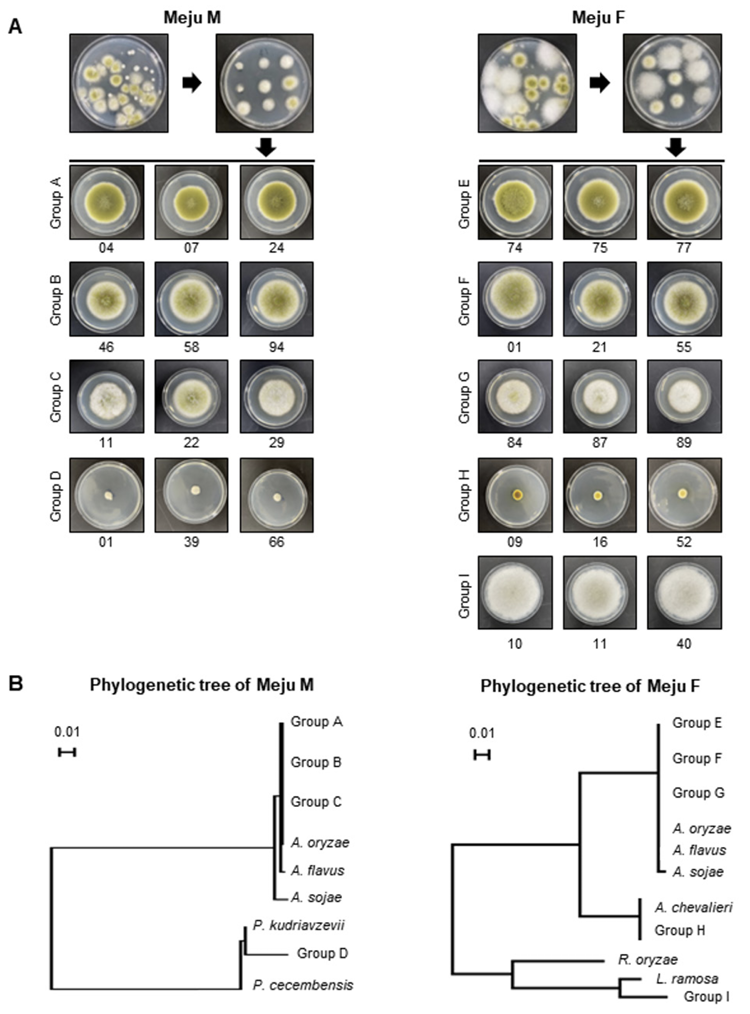
Fig. 2
Identification of fungal distribution using culture-based analysis. (A) The fungi isolated from Meju M (sample of the middle stage of meju fermentation) and Meju F (the final fermented meju products) samples were grouped by their morphological phenotypes. (B) Phylogenetic analyses of internal transcribed spacer (ITS) regions of the representative fungi in Meju M or Meju F samples. The sequences were first analyzed using the Tamura-Nei parameter distance calculation model, which were then used to construct the maximum likelihood tree with MEGA version 11. The bootstrap consensus tree is inferred through 1,000 replicates. Above the branches represent the bootstrap values (>50%) obtained for 1,000 replicates. The strain isolated in this study is indicated in bold. Bar, 0.02 substitutions per nucleotide position.
For further identification of the isolates, we selected three representative strains from each group, extracted the genomic DNA from each strain, and checked the ITS sequence. As shown in Fig. 2B, Groups A, B, C, E, F, and G belong to A. flavus/oryzae strains. Isolates in Groups D, H, and I belong to Pichia kudriavzevii, A. chevalieri, and Lichtheimia ramose, respectively.
Aflatoxin production ability
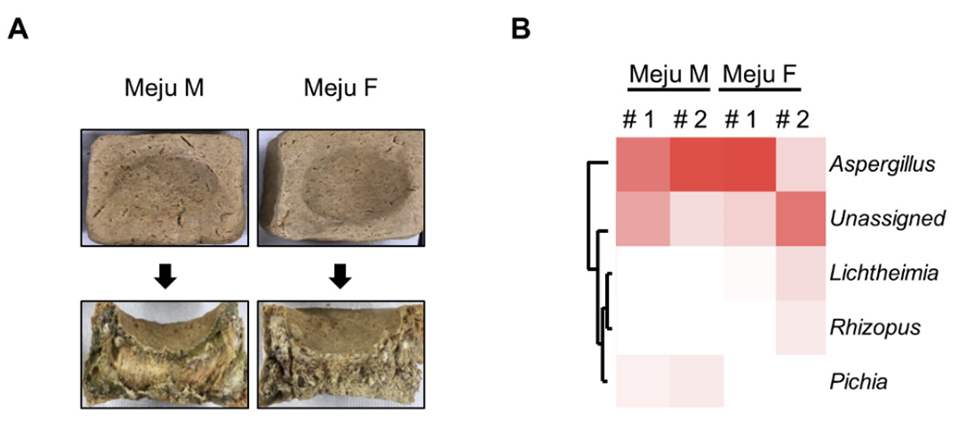
As mention earlier, A. flavus/oryzae strains are dominant, a result similar to that of previous studies [6,16]. Because A. flavus/oryzae strains might produce aflatoxins, it is important to confirm the production of aflatoxins. For this, we conducted TLC analysis and found that wild-type A. flavus, a positive control strain, produces aflatoxin B1, but all the test strains did not produce aflatoxin B1 (Fig. 3A).
A previous study proposed that the aflF-aflU interregion present in the aflatoxin biosynthetic gene cluster is an important region that can confirm the toxin production ability [16]. A. flavus, which produces aflatoxin B1, has the full length of aflF, but nontoxigenic A. flavus/oryzae strains have the partial length of aflF (Fig. 3B), so the functional aflF gene could not be transcribed. Therefore, we amplified the aflF-aflU interregion and checked the length of the PCR product. alfY was used as the positive control. As shown in Fig. 3C, the length of the aflF-aflU interregion in wild-type A. flavus was approximately 2.2 kB, but that in A. oryzae and all test strains was approximately 1.6 kB. Altogether, these results suggested that all the A. flavus/ oryzae strains isolated from Meju samples are nontoxigenic.
Fig. 3
Analysis of aflatoxin B1 production and aflatoxin gene cluster in fungal isolates. (A) Thin-layer chromatography (TLC) images representing the results of aflatoxin production in the representative strains. (B) The aflatoxin biosynthetic gene cluster shows difference in the length of aflatoxin-producing Aspergillus flavus and non-aflatoxin-producing A. oryzae. (C) Polymerase chain reaction (PCR) analysis of the aflF-aflU intergene region and aflE in the isolates. AFB1, aflatoxin B1; A. f , A. flavus; A. o, A. oryzae; M, marker.
Amylase and protease activities of isolates from meju
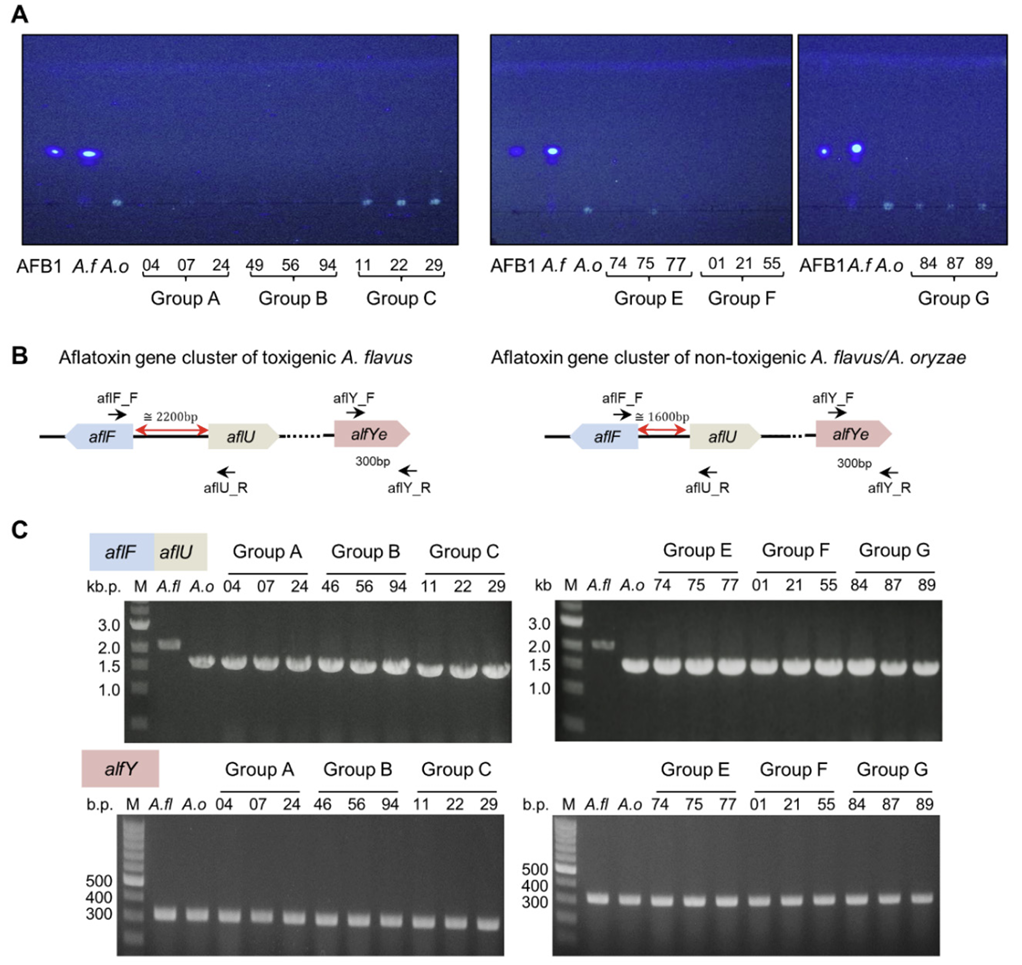
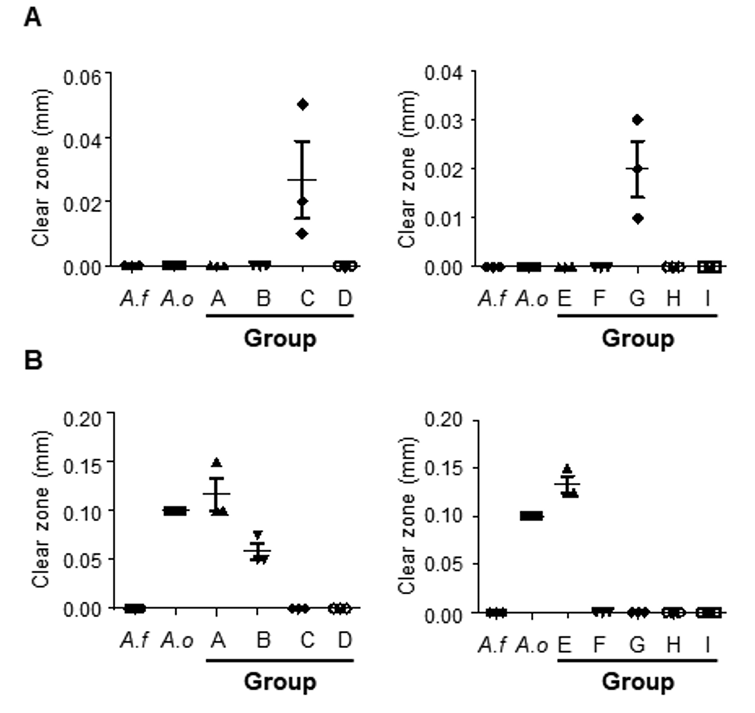
Enzymes produced by A. flavus/oryzae strains exert a major effect on various metabolites and flavors during meju fermentation. Especially, amylase and protease are key components for the decomposition of macromolecules in meju. Therefore, we evaluated the amylase and protease activities of representative strains from meju samples. Regarding amylase, we confirmed that Groups C and G had the largest clear zone radii, indicating that they can produce excessive amounts of amylase (Fig. 4A). Regarding proteases, Groups A and E showed the largest clear zone radii, indicating that the strains of these Groups A and E had relatively high proteolytic ability (Fig. 4B). These findings indicate that various fungi are present in fermented soybean bricks produced via traditional methods, and these isolates exhibit differences in enzyme activity.
Fig. 4
Enzyme activities of amylase and protease of the meju isolates. (A) Amylase activities of representative strains in each group are shown with the length of the clear zone. (B) Protease activities of representative strains in each group are shown with the length of the clear zone. A. f: Aspergillus flavus; A. o: A. oryzae.
DISCUSSION
In this study, fungi were isolated and identified from Meju M and Meju F samples obtained from traditionally fermented soybean paste producers in Gyeongsangbuk-do to identify fungal colonies and dominant fungi. Through metagenomic analysis and cultured-based analysis, we found that Aspergillus strains, especially A. flavus/oryzae, are dominant during meju fermentation. Pichia spp., including P. kudriavzevii, were found in Meju M samples but disappeared from the final product. The final meju product contained Rhizopus spp. and Lichtheimia spp.. The results of TLC and PCR showed that the representative strains lack some genes of the aflatoxin biosynthesis gene cluster and may not produce aflatoxin B1, suggesting that the fungi in meju do not produce mycotoxins. Moreover, the amylase and protease activities of these strains revealed that several A. flavus/oryzae strains produce amylase or protease. In conclusion, our results demonstrate the changes in fungal distribution during meju fermentation, and various fungi produce various enzymes such as protease and amylase to promote fermentation. Our findings also confirmed that the meju samples examined in this study were safe from aflatoxin-producing fungi.