INTRODUCTION
Fungi form different types of relationships with plants, including parasitic, saprophytic, and mutualistic relationships. Endophytic fungi form mutualistic relationship with plants, in which fungi inhabit plant tissues [1]. Unlike pathogenic fungi that cause plant diseases, endophytic fungi do not induce any disease symptoms in plants [2]. In contrast, they help protect plants from pathogens by involving their secondary metabolites [3,4]. Endophytic fungi reside in plant tissues, such as roots, stems, leaves, flowers, and fruits, and there are different endophytic fungal species based on the plant part where they reside [5,6].
Taean-gun, situated on the Taean Peninsula in the West Sea of Korea, features a distinctive rias coast marked by intricate and extensive coastline. The coastal area of Taean has a dune vegetation zone and was designated as Taean Costal National Park due to its significant conservation value. In previous studies, Kim et al. [7] investigated the distribution of higher fungi, and You et al. [8] reported endophytes isolated from the roots of halophytes in Taean-gun. Beyond the coastal environment, the region encompasses a Ramsar Convention wetland and low-height mountains, contributing to its diverse range of environments. Therefore, in this study, unrecorded endophytic fungi species were discovered from tissues of Korean oaks and Japanese cedar inhabit in the coast, wetlands, and forests of Taean-gun. Here, we report the morphological characteristics and phylogenetic analysis of three unrecorded endophytic fungal strains identified during the isolation process.
MATERIALS AND METHODS
Sample collection
Plant samples were collected between April and May 2022 from Taean-gun, Chungcheongnamdo, Korea. Pseudosasa japonica (Siebold & Zucc. ex Steud.) Makino ex Nakai is native to Anmyeondo island in Taean-gun (36°26'40.4"N 126°23'04.6"E), and Quercus serrata Murray were collected from Mangmisan Mountain (36°44'45.2"N 126°08'03.9"E) and Du-ung wetland (36°50'12.1"N 126°11'41.6"E). The plant samples were placed in polyethylene bags and transported to the laboratory within 24 hours.
Surface sterilization
The stems and roots of the plants were thoroughly cleaned with distilled water, then surface-sterilized by placing them in 1% NaClO solution for 1 minute and then in 70% EtOH for 2 min [9]. Surface-sterilized stems and roots were subdivided into appropriate sizes and inoculated onto potato dextrose agar (PDA; Difco Lab., Detroit, USA) medium.
Cultivation and morphological analysis
After inoculation, the PDA medium were cultured in a dark room at 25℃, with hyphal extension observed. When hyphae began to grow, the cultures were transferred to fresh PDA medium. The isolates were cultured on both PDA and malt extract agar (MEA; Kisan bio, Seoul, Korea) for 7 days, and their morphological characteristics were observed under an optical microscope (Axio Imager A1, Carl ZWISS, Overkochen, Germany).
Molecular analysis
Genomic DNA was extracted from the hyphae of morphologically classified fungal strains using a the HiGene Genomic DNA Prep Kit (BioFACT, Daejeon, Korea). For molecular identification, the internal transcribed spacer (ITS) region of rDNA was amplified using the primers ITS1F and ITS4 [10], the large subunit (LSU) region was amplified using the primers LR0R and LR16 [11], β-tubulin region was amplified using Bt2a and Bt2b [12], and the translation elongation factor 1-α region was amplified using the primers EF1-668F and EF1-1251R [13]. The PCR-amplified DNA was electrophoresed on a 1.5% agarose gel for 20 min to confirm the size of each DNA fragment. Thereafter, the DNA was subjected to sequence analysis (SolGent, Daejeon, Korea), and the similarities between DNA sequences were assessed using the BLAST tool on the National Center for Biotechnology Information (NCBI). For species identification and
Analyses of amylase and protease activities
The amylase activity was evaluated as described previously [16]. Each strain was inoculated into CM containing modified starch (20 g starch, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20 × nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Next, 2 mL Lugol solution (Sigma) was added to the plate to induce a blue–violet iodinestarch reaction. After removing the Logol solution, the width of the clear zone at the end of the mycelium was measured. For analyzing the activity of protease, each strain was inoculated into CM containing modified skim milk (20 g skim milk, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20× nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Then, the clear zone was measured.
phylogenetic analysis, sequences from two or three regions were concatenated, and a phylogenetic tree was constructed using the neighbor-joining method [14]. The identified endophytic fungal strains were deposited in the National Institute of Biological Resources (NIBR), and the DNA sequences used for the BLAST search and tree construction were registered with the NCBI.
RESULTS AND DISCUSSION
Geomyces asperulatus Sigler & J.W. Carmich., Mycotaxon 4 (2): 376 (1976) [MB#314398]
Morphological characteristics: The size of the colony cultured on PDA medium for 7 days ranged from 15.29-15.91 mm. The front of the colony was bright yellow, whereas the back was dark brown at the center and yellow-brown at the edges. The colony laid flat against the medium, and the medium around the colony was dyed black (Fig. 1A and 1G). The size of the colony cultured on MEA medium for 7 days ranged from 13.91 to 14.56 mm. The front was bright cream-colored, and the back was beige. The colony was flat against the medium and the edges form irregular but smooth curves (Fig. 1B and 1H). Conidiophores branch out from the tip of the mycelial growth, forming cylindrical conidia (Fig. 1M). The conidia were transparent with a hint of yellow and gave a rough, sandpaper-like feel on their surface. The conidia lacked septa, and their size was approximately (2.68-) 3.15 (-3.86)×(2.33-) 2.69 (-3.07) μm (n=50) (Fig. 1N).
Fig. 1
Cultural characteristics of three endophytic fungal strains. Colonies of Geomyces asperulatus KNUE 22T049 grown for 7 days on potato dextrose agar (PDA) (A, G), malt extract agar (MEA) (B, H), conidiophore (M) and conidia (N); Colonies of Leptoxyphium fumago KNUE 22N512 grown for 7 days on PDA (C, I), MEA (D, J), immature conidia (O) and mature conidia (P); Colonies of Tubakia oblongispora KNUE 22N2108 grown for 7 days on PDA (E, K), MEA (F, L), conidiophore (Q) and conidia (R) (scale bars=10 μm).
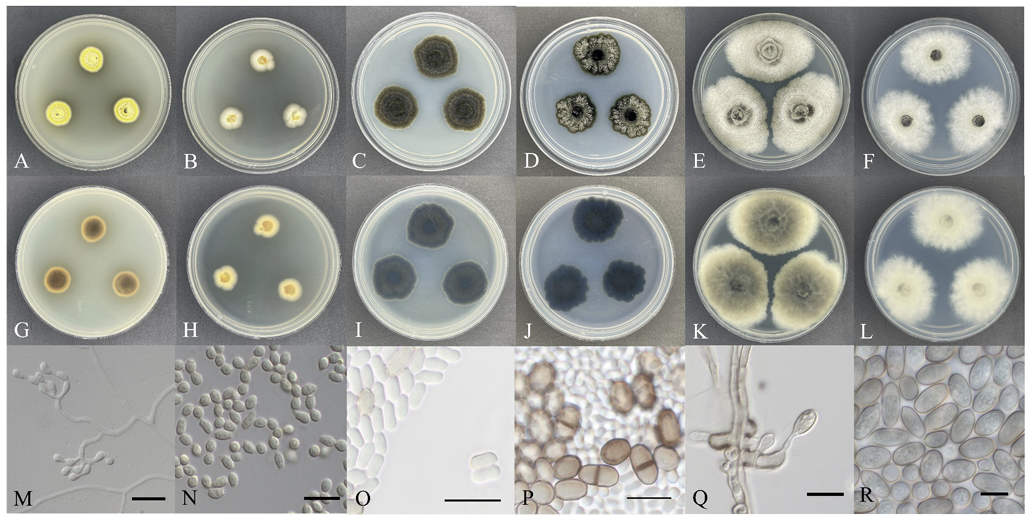
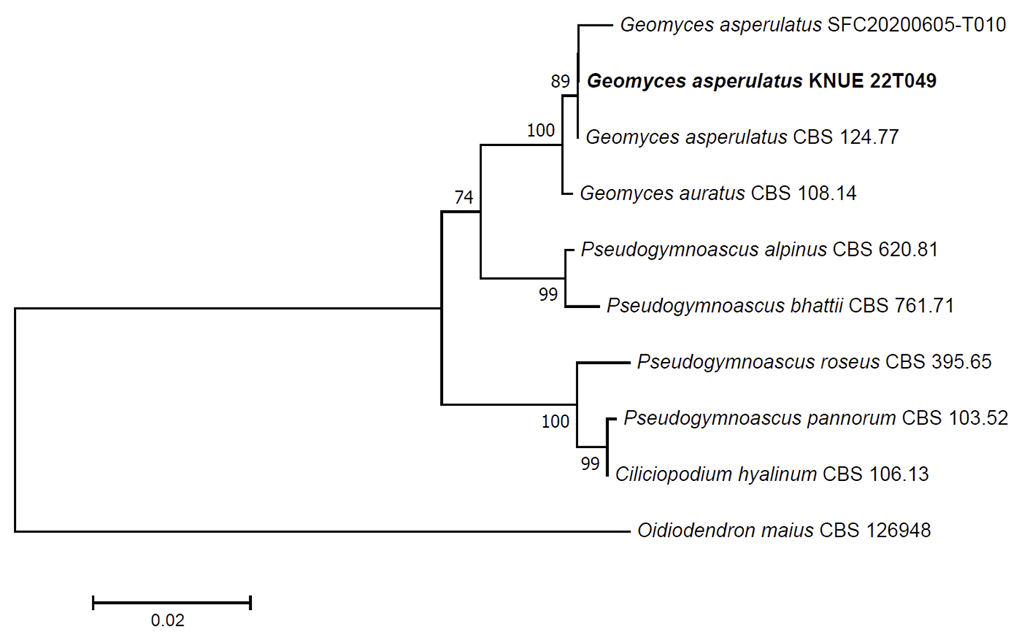
Fig. 2
Neighbor-joining phylogenetic tree of based on a concatenated alignment of internal transcribed spacer (ITS), large subunit (LSU) sequences. Oidiodendron maius was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in a bold.
Specimen examined: Taean-gun, Chungcheongnam-do, Korea, 36°44'45.2"N 126°08'03.9"E, May 19, 2022, Geomyces asperulatus, isolated from root of Quercus serrata, strain KNUE 22T049, NIBRFGC000510027, GenBank No. OR054224 (ITS) and OR054226 (LSU).
Notes: G. asperulatus was first reported in 1976, with its species epithet derived from “Asperulate”, meaning “Easily roughened” [15,16]. They were characterized by cylindrical conidia that formed and inflated at the ends of the conidiophores, branching out like tree branches (Table 1) [17]. Strain KNUE 22T049, isolated from the root of Q. serrata, was confirmed to contain conidiophores and conidia consistent with the characteristics recorded in the original description. G. asperulatus grows well at low temperatures around 10℃ [18], and has been recorded as an endophyte isolated from the fruit of grape (Vitis vinifera), suggesting a potential contribution to the formation of grape polyphenols [19]. Molecular analysis revealed that the ITS region had 99.64% similarity with G. asperultus OW982393.1, and the LSU region had 100% similarity with G. asperultus MH861038.1. The NJ phylogenetic tree constructed using sequences of ITS and LSU regions revealed that the strain KNUE 22T049 formed with a monophyletic clade with the strains, G. asperulatus CBS 124.77 and SFC20200605-T010 (Fig. 2).
Leptoxyphium fumago (Woron.) Crous, Fungal Systematics and Evolution 6: 198 (2020) [MB#835082]
Morphological characteristics: The size of the colony cultured on PDA medium for 7 days ranged from 26.05 to 27.46 mm, with both the front and back exhibiting a gray-brown color in the center and a dark brown at the edges. The colony was slightly raised from the medium, and radiated towards the edges (Fig. 1C and 1I). The size of the colony cultured on MEA medium for 7 days ranged from 22.95 to 27.66 mm, with the front appearing grey-white and the back appearing slate-blue. The colony laid flat against the medium and the edges had irregular wave-like shapes (Fig. 1D and 1J). Conidiophores formation was not observed; however, both immature and mature conidia were observed. The immature conidia were transparent cylindrical in shape, lacked septa, and measured about (3.74-) 4.44 (-5.52)×(1.79-) 2.15 (-2.74) μm (n=50) (Fig. 1O). The mature conidia were pigmented pale brown and were sometimes divided by a layer of septa. Their size was approximately (6.45-) 7.95 (-10.94)×(3.10-) 4.50 (-5.84) μm (n=50) (Fig. 1P).
Specimen examined: Taean-gun, Chungcheongnam-do, Korea, 36°26'40.4"N 126°23'04.6"E, April 8, 2022, Leptoxyphium fumago, isolated from stem of Pseudosasa japonica, strain KNUE 22N512, NIBRFGC000510029, GenBank No. OR054223 (ITS) and OR054222 (LSU).
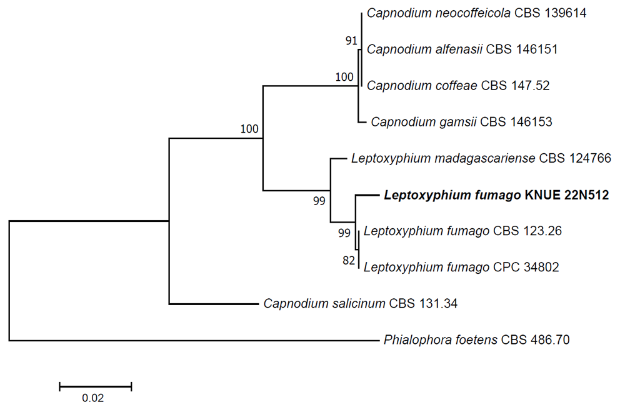
Notes: The first recorded name for L. fumago was Fumago vagans, which was reported by Persoon in 1822. It has also been placed in the genera Cladosporium and Caldariomyces. The current species name was proposed by Srivast in 1982 and reconfirmed in 2020 [20]. Immature conidia are transparent and lack septa; however, septa can be observed in mature spores, and their size increases [21]. The conidia morphology of strain KNUE 22N512, which was isolated from the stems of P. japonia in this study, was consistent with these characteristics. L. fumago has been reported to parasitize the exudates of insects living on plant leaves [21], and has also been isolated from the leaves of Citrus spp. [20]. It secrets the enzyme chloroperoxidase when grown in glucose-malt medium [22]. Sequence analysis confirmed a 99.19% match in the ITS region with L. fumago MT223811 and a 99.67% match in the LSU region with L. fumago MT223906. According to the NJ phylogenetic tree constructed using sequences of ITS and LSU regions, the strain KNUE 22N512 distinctly clustered with the strains, L. fumago CBS 123.26 and CPC 34802 (Fig. 3).
Fig. 3
Neighbor-joining phylogenetic tree of based on a concatenated alignment of internal transcribed spacer (ITS) and large subunit (LSU) sequences. Phialophora foetens was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strains isolated in this study are in bold.
Tubakia oblongispora C. Nakash., Fungal Systematics and Evolution 1: 86 (2018) [MB#823665]
Morphological characteristics: The size of the fungal colony cultured on PDA medium for 7 days was approximately 37.83-39.74 mm, the front was dark beige, and the back was dark grey. The colony was slightly fused in the medium, and the wave pattern spread from the center of the colony to the edge (Fig. 1E and 1K). The size of the colony cultured on the MEA medium for 7 days was approximately 32.29-34.25 mm, and both the front and back were light beige. The colony was flat on the medium, and the aerial mycelia spreading towards the edge and looked similar to cotton wool (Fig. 1F and 1L). Oval conidiophores formed along the direction of mycelial growth, and conidia were generated diagonally from the conidiophores (Fig. 1Q). The semi-transparent conidia shone brown on the outer wall, were elliptical or oblong in shape, and their size was approximately (10.80-) 12.43 (-14.06)×(5.74-) 6.61 (-7.96) μm (n=50) (Fig. 1R).
Specimen examined: Taean-gun, Chungcheongnam-do, Korea, 36°50'12.1"N 126°11'41.6"E, May 20, 2022, Tubakia oblongispora, isolated from branch of Quercus serrata, strain KNUE 22N2108, EKFTFGC000000062, GenBank No. OR054227 (ITS), OR059182 (β-tubulin, TUB) and OR059183 (translation elongation factor 1-α, TEF)
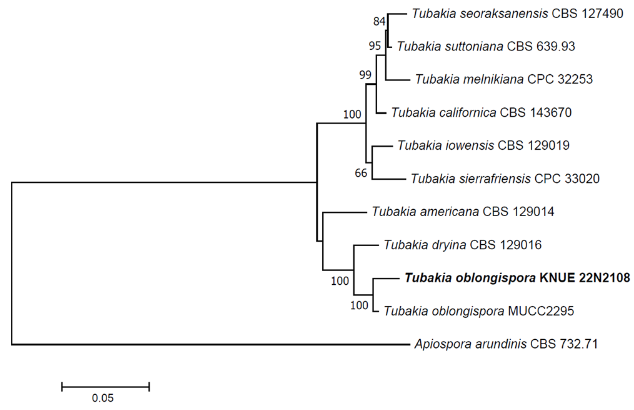
Notes: T. oblongispora was first reported in 2018. It was morphologically and phylogenetically similar to Tubakia dryina, but was recorded as a new species due to morphological differences in the formation of oblong spores [23]. It formed ellipsoidal, conical, or cylindrical spores [23], and the spore-bearing hyphae and spores of the strain KNUE 22N2108, isolated from the stems of Q. serrata in this study, matched the characteristics and sizes recorded in the original description. T. oblongispora has also been recorded in Japan as an endophyte from the leaves of Q. serrata [23]. Sequence analysis confirmed that the ITS region matched 99.09% with T. oblongispora NR159055, the TUB region matched 99.40% with T. oblongispora MG592178, and the TEF region matched 100% with T. oblongispora MG592084. The NJ phylogenetic tree constructed using sequences of ITS, TUB and TEF regions revealed that the strain KNUE 22N2108 formed with a monophyletic clade with the strains, T. oblongispora MUCC2295 (Fig. 4).
Fig. 4
Neighbor-joining phylogenetic tree of based on a concatenated alignment of internal transcribed spacer (ITS), β-tubulin (TUB) and translation elongation factor 1-α (TEF) DNA sequences. Apiospora arundinis was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strains isolated in this study are in bold.