Glechoma longituba (Nakai) Kuprian. (syn. Glechoma hederacea var. longituba Nakai) of the Lamiaceae family, known as long tube ground ivy, is native to Vietnam, China, Korea, and the Russian Far East (https:// powo.science.kew.org/). This plant has been used for medicinal purposes in Asia since long [1,2]. In Korea, these plants are dried and marketed as herbal tea [3].
Powdery mildew infections of Glechoma L. species are known to be associated with Golovinomyces biocellatus (Ehrenb.) V. P. Heluta [4]. In Korea, this pathogen is associated with Agastache rugosa Kuntza, Meehania urticifolia Makino, Mentha suaveolens Ehrh., Monarda citriodora Cerv. ex Lag., M. didyma L., and Rosmarinus officinalis L. from the Lamiaceae family [5-10]. Powdery mildew from the Golovinomyces genus associated with host plants of the Lamiaceae family are known to be a part of the G. biocellatus complex [11]. Recent molecular phylogenetic analyses have resolved the taxonomic composition of this complex by categorizing it into several new combinations, including Golovinomyces monardae (G.S. Nagy) M. Scholler, U. Braun & Anke Schmidt, G. neosalviae M. Scholler, U. Braun & Anke Schmidt, and G. salviae (Jacz.) M. Scholler, U. Braun & Anke Schmidt [12]. Currently, Golovinomyces bicellatus sensu stricto is considered to be a pathogen of Glechoma and Lycopus L. species.
During routine plant disease surveys in 2022, G. longituba plants were found to be infected with powdery mildew in Jeongeup, Jeonju, Buan, Jinan, and Wanju, Korea. Subsequently, six samples were deposited in the Mycological Herbarium of Korea University, namely KUS-F32257 (June 15, 2021, Hongcheon), F32837 (May 20, 2022, Jeongeup), F32916 (June 7, 2022, Jeonju), F32934 (June 10, 2022, Wanju), F32977 (June 20, 2022, Buan), and F33540 (November 13, 2022, Jinan).
For morphological examination, a small piece of the fungal colony was scraped off the infected leaves and mounted on a drop of distilled water on a glass slide. Subsequently, they were examined under an optical microscope using bright-field and differential interference contrast (DIC). At least 30 measurements of each structure were taken using an Olympus BX51 microscope (Olympus, Tokyo, Japan) at 40× and 100× magnifications, and a Zeiss AX10 microscope equipped with an AxioCam MRc5 (Carl Zeiss, Göttingen, Germany) was used for capturing the photographs.
Powdery mildew colonies first appeared as circular to irregular white patches but soon progressed to abundant hyphal growth on both sides of the leaves and young stems (Fig. 1A and B). Severe infection caused the withering and premature senescence of leaves in plants, reducing their vigor. In powdery mildew, the hyphae were septate, branched, and 4-9 µm wide, and the hyphal appressoria were nipple-shaped. Conidiophores measured 120-200×10-12 μm and produced 1-4 immature conidia in chains with a sinuate outline, followed by 2-3 cells (Fig. 1C-E). The foot-cells of conidiophores were straight, cylindrical, and 3555 μm long. Conidia were hyaline, were ellipsoid to barrel-shaped, measured 22-30×19-23 μm (length/ width ratio 1.1-1.6), lacked distinct fibrosin bodies, and produced germ tubes at the subterminal position (Fig. 1F). Chasmothecia were not observed. Short conidiophores and a low length/width ratio of conidia are typical characteristics indicating the asexual state of Golovinomyces salviae [12].
MATERIALS AND METHODS
Sample collection
Plant samples were collected between April and May 2022 from Taean-gun, Chungcheongnamdo, Korea. Pseudosasa japonica (Siebold & Zucc. ex Steud.) Makino ex Nakai is native to Anmyeondo island in Taean-gun (36°26'40.4"N 126°23'04.6"E), and Quercus serrata Murray were collected from Mangmisan Mountain (36°44'45.2"N 126°08'03.9"E) and Du-ung wetland (36°50'12.1"N 126°11'41.6"E). The plant samples were placed in polyethylene bags and transported to the laboratory within 24 hours.
Surface sterilization
The stems and roots of the plants were thoroughly cleaned with distilled water, then surface-sterilized by placing them in 1% NaClO solution for 1 minute and then in 70% EtOH for 2 min [9]. Surface-sterilized stems and roots were subdivided into appropriate sizes and inoculated onto potato dextrose agar (PDA; Difco Lab., Detroit, USA) medium.
Cultivation and morphological analysis
After inoculation, the PDA medium were cultured in a dark room at 25℃, with hyphal extension observed. When hyphae began to grow, the cultures were transferred to fresh PDA medium. The isolates were cultured on both PDA and malt extract agar (MEA; Kisan bio, Seoul, Korea) for 7 days, and their morphological characteristics were observed under an optical microscope (Axio Imager A1, Carl ZWISS, Overkochen, Germany).
Molecular analysis
Genomic DNA was extracted from the hyphae of morphologically classified fungal strains using a the HiGene Genomic DNA Prep Kit (BioFACT, Daejeon, Korea). For molecular identification, the internal transcribed spacer (ITS) region of rDNA was amplified using the primers ITS1F and ITS4 [10], the large subunit (LSU) region was amplified using the primers LR0R and LR16 [11], β-tubulin region was amplified using Bt2a and Bt2b [12], and the translation elongation factor 1-α region was amplified using the primers EF1-668F and EF1-1251R [13]. The PCR-amplified DNA was electrophoresed on a 1.5% agarose gel for 20 min to confirm the size of each DNA fragment. Thereafter, the DNA was subjected to sequence analysis (SolGent, Daejeon, Korea), and the similarities between DNA sequences were assessed using the BLAST tool on the National Center for Biotechnology Information (NCBI). For species identification and
Analyses of amylase and protease activities
The amylase activity was evaluated as described previously [16]. Each strain was inoculated into CM containing modified starch (20 g starch, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20 × nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Next, 2 mL Lugol solution (Sigma) was added to the plate to induce a blue–violet iodinestarch reaction. After removing the Logol solution, the width of the clear zone at the end of the mycelium was measured. For analyzing the activity of protease, each strain was inoculated into CM containing modified skim milk (20 g skim milk, 1 g yeast extract, 1.5 g casamino acids, 2 g peptone, 50 mL of 20× nitrate salt solution, 1 mL of 1,000× trace element, and 15 g agar per 1 L), and the plate was cultured for 4 days at 30℃. Then, the clear zone was measured.
phylogenetic analysis, sequences from two or three regions were concatenated, and a phylogenetic tree was constructed using the neighbor-joining method [14]. The identified endophytic fungal strains were deposited in the National Institute of Biological Resources (NIBR), and the DNA sequences used for the BLAST search and tree construction were registered with the NCBI.
RESULTS AND DISCUSSION
Geomyces asperulatus Sigler & J.W. Carmich., Mycotaxon 4 (2): 376 (1976) [MB#314398]
Morphological characteristics: The size of the colony cultured on PDA medium for 7 days ranged from 15.29-15.91 mm. The front of the colony was bright yellow, whereas the back was dark brown at the center and yellow-brown at the edges. The colony laid flat against the medium, and the medium around the colony was dyed black (Fig. 1A and 1G). The size of the colony cultured on MEA medium for 7 days ranged from 13.91 to 14.56 mm. The front was bright cream-colored, and the back was beige. The colony was flat against the medium and the edges form irregular but smooth curves (Fig. 1B and 1H). Conidiophores branch out from the tip of the mycelial growth, forming cylindrical conidia (Fig. 1M). The conidia were transparent with a hint of yellow and gave a rough, sandpaper-like feel on their surface. The conidia lacked septa, and their size was approximately (2.68-) 3.15 (-3.86)×(2.33-) 2.69 (-3.07) μm (n=50) (Fig. 1N).
Fig. 1
Powdery mildew caused by Golovinomyces salviae on Glechoma longituba. A, B: Symptoms on the leaves. C-E: Conidiophores. F: Conidia.

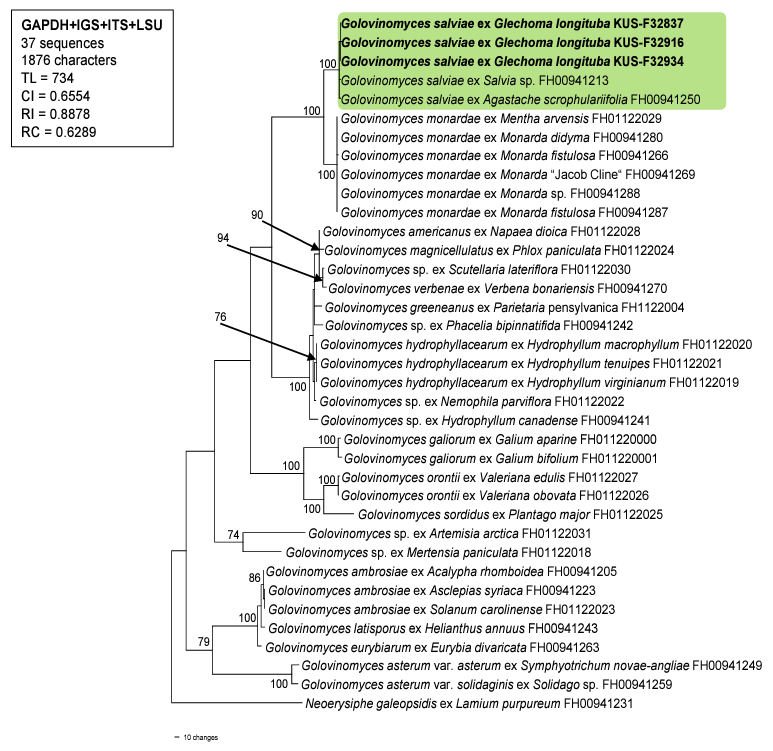
For molecular phylogenetic analyses, genomic DNA was extracted from the mycelia of three samples (i.e., KUS-F32837, F32916, and F32934) using MaglistoTM 5M kits (Bioneer, Daejeon, Korea), following the manufacturer’s instructions. The nucleotide sequences of the internal transcribed spacer (ITS) and intergeneric spacer (IGS) regions as well as the large subunit (LSU) of the rDNA, and glyceraldehyde3-phosphate dehydrogenase (GAPDH) gene were amplified and sequenced using the primer pairs ITS1/ PM6, IGS-12a/NS1R, PM3/TW14, and PMGAPDH1/PMGAPDH3R, respectively [13,14]. Newly obtained sequences were assembled and deposited in the NCBI database under the accession numbers OP872648, OP872652, and OP877276 for ITS; OP872651, OP872653, and OP872655 for LSU; OQ281744-OQ281746 for IGS; and OQ281747-OQ281749 for GAPDH. These sequences were then combined in a multigene dataset of GAPDH+IGS+ITS+LSU with 33 Golovinomyces sequences retrieved from GenBank, according to method described by Bradshaw et al. [14]. Following this, they were aligned using the MUSCLE command implemented in MEGA 11 [15]. Neoerysiphe galeopsidis (DC.) U. Braun was selected as the outgroup. A phylogenetic tree was generated based on the maximum parsimony (MP) method in PAUP* 4.0a, with the heuristic search option using the ʻtree-bisection-reconstruction’ (TBR) algorithm. All sites were treated as unordered and unweighted, and gaps were treated as missing data [16]. The robustness of the internal branches was evaluated using bootstrap (BS) analysis with 1,000 replications. The tree scores calculate were tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC).
The resulting sequences of ITS were 100% identical to those of G. salviae (AB769437 and LC100001 etc.) and G. biocellatus (AB307669), whereas sequences of LSU showed 99.87% similarity with G. salviae (LC076803 and LC100001). The GAPDH and IGS results showed 99.6% and 99.4% similarity to G. salviae (ON075654 and ON075655), respectively. In total, 37 sequences and 1,876 characters were used in the MP analysis. Among the 1,876 characters, 1,491 (79.4%) were constant, 281 (14.9%) were parsimonyinformative, and 104 (5.54%) were variable and parsimony-uninformative. In the consensus tree (Fig. 2), the sequences obtained from the three Korean isolates were clustered in a distinct clade with the sequences of G. salviae, and the branch was supported with a 100% bootstrap value.
Glechoma species belong to the Mentheae tribe of the Lamiaceae family. According to a study on Golovinomyces biocellatus complex conducted by Schoeller et al. [12], most host plant species from this tribe, such as Mentha L., Monarda L., Origanum L., and Thymus L. spp., are infected by Golovinomyces monardae. In contrast, Glechoma hederacea L. has been reported to be a target host plant for G. biocellatus sensu stricto. However, G. salviae was reported as a new combination that was excluded from the biocellatus complex, owing to its pathogenicity to Salvia spp. (tribe Mentheae) in Europe. Recently, Agastache scrophulariifolia (Wild.) Kuntza, from the Mentheae tribe, has been recorded as the host plant of this fungus in the USA [14]. Our research suggests that the host-plant range of G. salviae is increasing, and the target host plants of powdery mildew pathogen (from the Mentheae tribe) are not confined to one species, but includes several Golovinomyces species. To the best of our knowledge, this is the first report of powdery mildew caused by G. salviae on G. longituba, and the first report of powdery mildew in Korea.
Fig. 2
A phylogenetic tree of Golovinomyces species is generated from a combined multigene dataset of GAPDH+IGS+ITS+LSU sequences based on the maximum parsimony method. Neoerysiphe galeopsidis was used as an outgroup. The newly obtained sequences are shown in bold. Bootstrap values (>70%) are indicated on related branches. Tree scores such as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC) are given in the box. GAPDH, glyceraldehyde-3phosphate dehydrogenase; IGS, intergeneric spacer; ITS, internal transcribed spacer; LSU, large subunit.
Specimen examined: Taean-gun, Chungcheongnam-do, Korea, 36°44'45.2"N 126°08'03.9"E, May 19, 2022, Geomyces asperulatus, isolated from root of Quercus serrata, strain KNUE 22T049, NIBRFGC000510027, GenBank No. OR054224 (ITS) and OR054226 (LSU).


