INTRODUCTION
Yeasts are eukaryotic microorganisms that play an important role in fermentation and have been isolated from various natural environments, including food [1]. Since 2000, towing to the global increase in need for research to protect biological resources, studies on wild yeasts isolated from various natural environments have been actively conducted in Korea [2]. Yeasts have various biological activities and are used to produce vitamins and amino acids. Additionally, they have recently been reported as probiotic microorganisms. As a result, industrial applications of yeast are rapidly increasing [3].
However, many yeast species that are widely used in research and industry are not included in the National Species List of Korea (NSLK), an important reference material for claiming rights to biological resources and the importance of which, has increased with the adoption of the Nagoya Protocol [3]. Therefore, this study aimed to analyze the taxonomic, morphological, and physiological characteristics of seven yeast strains not yet included in the NSLK, namely Hyphopichia burtonii, Starmerella sorbosivorans, Cyberlindnera mycetangii, Cutaneotrichosporon oleaginosum, Nakazawaea ernobii, Pichia kudriavzevii, and Schizosaccharomyces japonicus, for their inclusion in the NSLK.
MATERIALS AND METHODS
Species and strain selection
Fifty yeast strains were isolated from various regions and materials in Korea, including nuruk, foxtail weeds, and pine trees. Among these, seven yeast strains that were not recorded in the NSLK were selected for taxonomic evaluation. Samples originating from different locations were collected in sterilized tubes, crushed using a mortar and pestle, and soaked overnight in 5 mL of saline solution to obtain a suspension. Part of the suspension was plated onto YPD agar containing ampicillin, DRBC agar, and DG18 agar. Yeast colonies growing after 48 h at 25℃ were picked and isolated as a single colony for further identification [1].
DNA isolation, amplification, and phylogenetic analysis
The isolated yeast strains were identified by analyzing their molecular biological relationships with yeast strains registered in the NCBI database using the nucleotide sequence of the D1/D2-26S rRNA region. The D1/D2-26S rRNA region was amplified via PCR as follows: the primers NL-1 (5'-GCATATCAAT AAGCGGAGGAAAAG-3') and NL-4 (5'-GGTCCGTGTTTCAAGACGG-3') were used with a predenaturation step at 95℃ for 5 min, followed by 30 cycles of denaturation at 95℃ for 30 s, annealing at 55℃ for 30 s, and extension at 72℃ for 30 s, as well as a post-extension step for 10 min at 72℃. The D1/ D2 region sequences of the isolated strains were deposited in GenBank (https://www.ncbi.nlm.nih.gov/ genbank/).
Phylogenetic analysis was performed using the D1/D2-26S rRNA region sequence. To this end, the neighbor-joining method based on the Tamura-Nei model was used to construct a phylogenetic tree using MEGA X software. Bootstrap analysis was performed 1,000 times. Taxonomic classification was performed using the type strains of a species within a genus or family [4].
Carbon source assimilation and oxidation
The physiological characteristics, including carbon source assimilation and oxidation and the morphology of each yeast strain were analyzed. Carbon source assimilation and oxidation experiments were performed using YT microplates (Biolog, Hayward, CA, USA). Carbon source assimilation was measured by turbidity, while oxidation was determined by color change. All measurements were performed using a microplate reader according to the manufacturer’s recommendations (Biolog).
Morphology
Cell morphology was examined by observing cells cultured on YM agar plates for 3 d at 25℃ using a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan). Colony formation was observed by inoculating a single colony and growing for 7 d at 25℃. Hyphae and pseudohyphae formation were examined using Dalmau plates on cornmeal agar for 14 d at 25℃ [5].
RESULTS AND DISCUSSION
In this study, we conducted a taxonomic evaluation of seven yeast strains that have not been previously documented in Korea: six ascomycetous yeasts (Hyphopichia burtonii, Starmerella sorbosivorans, Cyberlindnera mycetangii, Nakazawaea ernobii, Pichia kudriavzevii, and Schizosaccharomyces japonicus) and one basidiomycetous yeast (Cutaneotrichosporon oleaginosum). The specific origins of these seven strains are listed in Table 1. Phylogenetic analyses were performed by determining and comparing the D1/ D2-26S rRNA region of the large subunit ribosomal ribonucleic acid (LSU rRNA) gene of the selected strains, which were evaluated and grouped with closely related species obtained from the National Center for Biotechnology Information (NCBI) database.
A yeast strain obtained from Nuruk formed a single clade with the type strain of H. burtonii CBS 2352 (Fig. 1A). Similarly, a strain isolated from Rubus coreanus formed a monophyletic clade with the type strain S. sorbosivorans CBS 8768 (Fig. 2A). A yeast strain isolated from Pinus densiflora bark developed a monophyletic clade with the type strains of Cy. mycetangii NRRL Y-6843 and CBS 8675 (Fig. 3A). A strain isolated from the bark of a deciduous tree in Busan formed a clade distinct from the type strain of Cu. oleaginosum ATCC 20509 (Fig. 4A). Conversely, the strain isolated from Camellia sasanqua was categorized into a single clade with the type strain N. ernobii CBS 1737 (Fig. 5A). A strain isolated from Hydrangea serrata formed a distinct clade with type strains P. kudriavzevii NRRL Y 5396 and CBS 5147 (Fig. 6A). Finally, a strain isolated from the bark of a deciduous tree in Yongin was grouped in a monophyletic clade with the type strain S. japonicus NRRL Y 1361 (Fig. 7A). Further analysis of the cell morphology and physiological characteristics of these strains provided supportive data for including these seven species on the NSLK list.
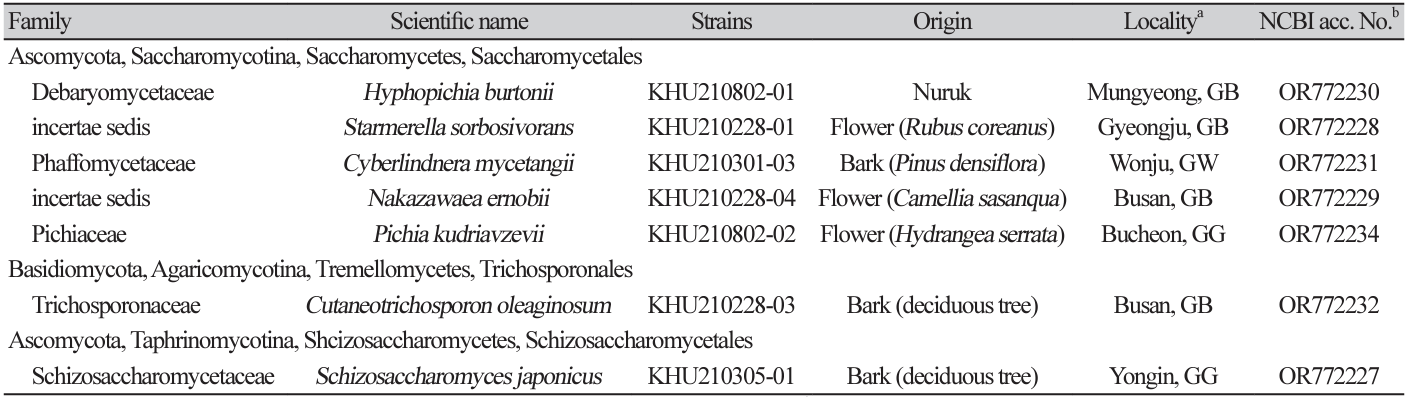
Fig. 1
Phylogenetic tree and morphological characteristics of Hyphopichia burtonii. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of H. burtonii strains isolated from Korea. Bold means representative strain. B-E. Morphology of H. burtonii KHU210802-01. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. hyphae formed on YM agar 7 days at 25℃. E. Septate hyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
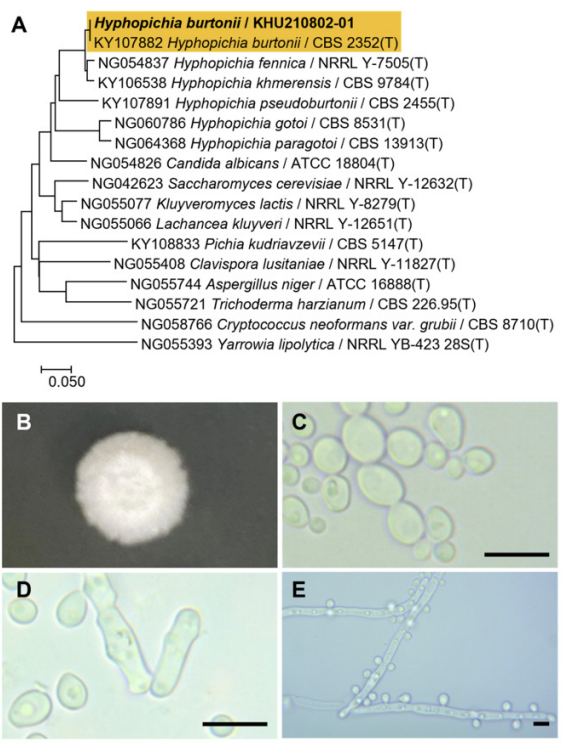
Fig. 2
Phylogenetic tree and morphological characteristics of Starmerella sorbosivorans. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of S. sorbosivorans strains isolated from Korea. Bold means representative strain. B-E. Morphology of S. sorbosivorans KHU210228-01. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Budding cells on YM agar 7 days at 25℃. E. Pseudohyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
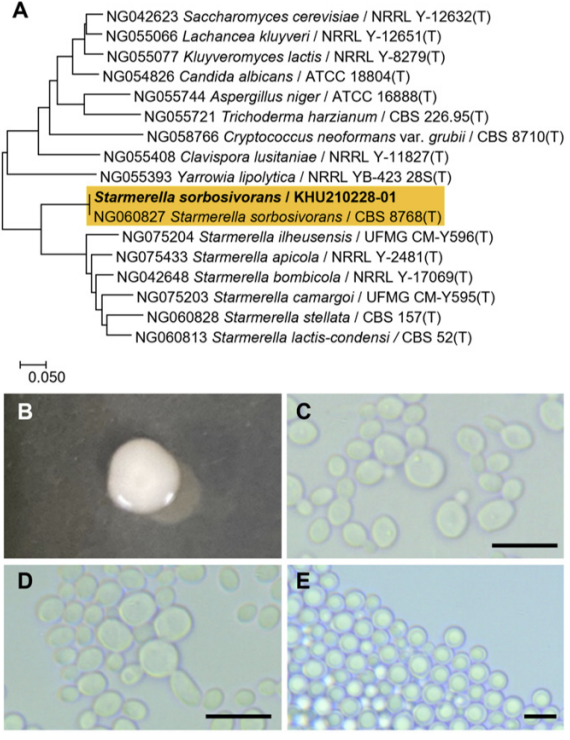
Fig. 3
Phylogenetic tree and morphological characteristics of Cyberlindnera mycetangii. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of C. mycetangii strains isolated from Korea. Bold means representative strain. B-E. Morphology of C. mycetangii KHU210301-03. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Hyphae formed on YM agar 7 days at 25℃. E. Septate hyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
Fig. 4
Phylogenetic tree and morphological characteristics of Cutaneotrichosporon oleaginosum. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of C. oleaginosum strains isolated from Korea. Bold means representative strain. B-E. Morphology of C. oleaginosum KHU210228-03. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Budding cells on YM agar 7 days at 25℃. E. Pseudohyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
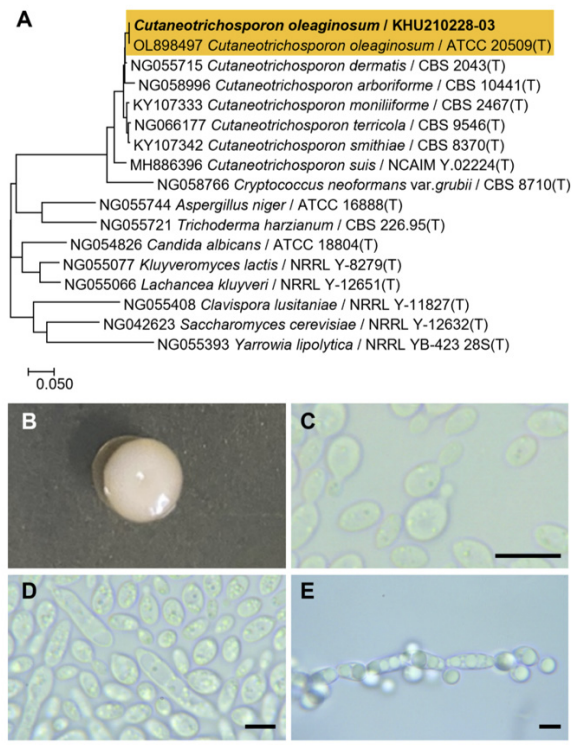
Fig. 5
Phylogenetic tree and morphological characteristics of Nakazawaea ernobii. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of N. ernobii strains isolated from Korea. Bold means representative strain. B-E. Morphology of N. ernobii KHU210228-04. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Budding cells on YM agar 7 days at 25℃. E. Pseudohyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
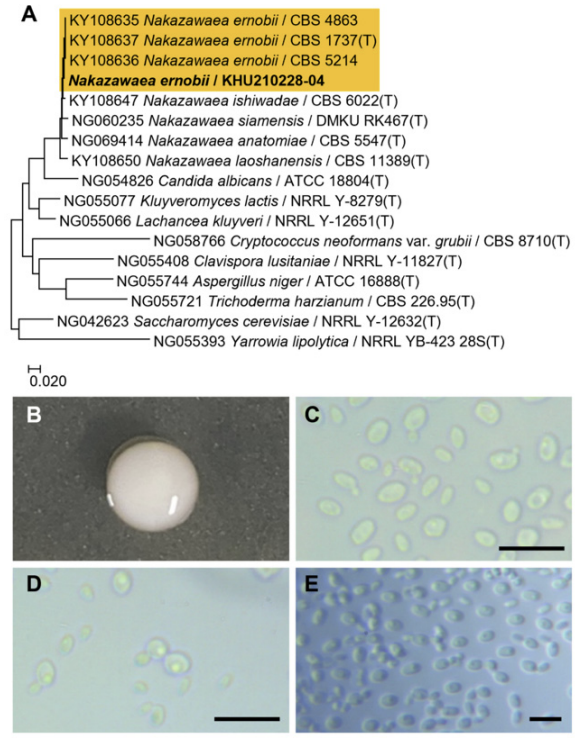
Fig. 6
Phylogenetic tree and morphological characteristics of Pichia kudriazevii. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of P. kudriazevii strains isolated from Korea. Bold means representative strain. B-E. Morphology of P. kudriazevii KHU210802-02. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. hyphae formed on YM agar 7 days at 25℃. E. hyphae formed on Dalmau plate with cornmeal agar for 2 weeks. Bars, 10 µm.
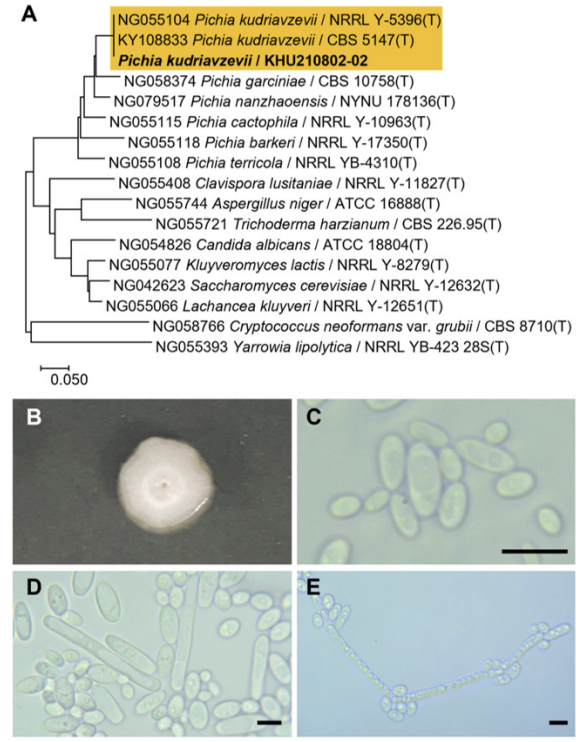
Fig. 7
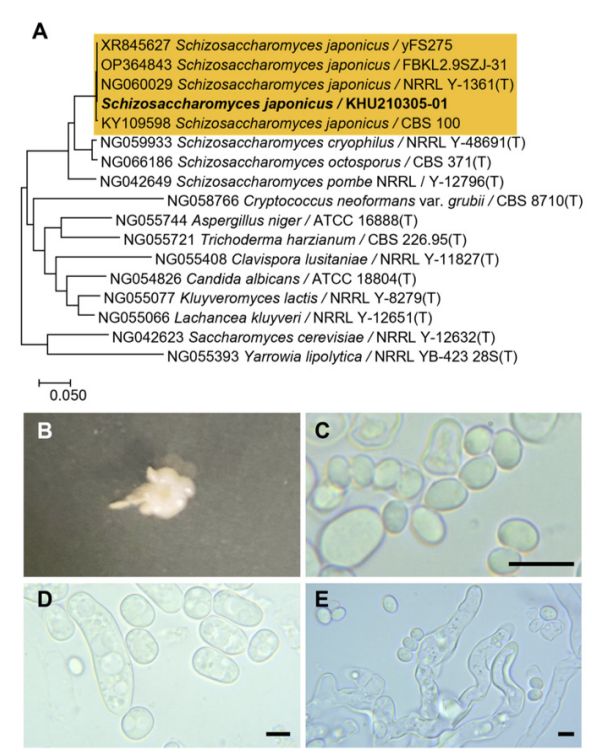
Phylogenetic tree and morphological characteristics of Schizosaccharomyces japonicus. A. Phylogenetic tree drawn from neighbor-joining analysis based on the D1/D2-26S domains of the large subunit (LSU) rRNA sequences, showing positions of S. japonicus strains isolated from Korea. Bold means representative strain. B-E. Morphology of S. japonicus KHU210305-01. B. Colony on YM agar 3 days at 25℃. C. Budding cells on YM agar 3 days at 25℃. D. Hyphae formed on YM agar 7 days at 25℃. E. hyphae formed on Dalma.
SPECIES DESCRIPTION
Hyphopichia burtonii (Boidin, Pignal, Lehodey, Vey & Abadie) Arx & Van der Walt, Antonie van Leeuwenhoek 42: 309-314, 1976
Hyphopichia is a fungal genus that belongs to the Saccharomycetales order. The genus Hyphopichia was originally introduced by Arx and van der Walt [6] to include the mycelial yeast Pichia burtonii. This species is heterothallic and forms asci by conjugating with yeast cells. In addition, this genus forms septate hyphae and conidia borne by the denticles. Arx and van der Walt [6] excluded P. burtonii from the genus Pichia based on the latter characteristic and from the genus Saccharomycopsis based on the former characteristic. However, the reclassification of P. burtonii was not accepted by Kurtzman and Fell [7] as other yeasts and dimorphic Euascomycetes share these characteristics.
The colonies appeared white to ivory in color, while the cells were ovoid to elongated with a size of 4.65.8×5.9-8.7 μm and occurred either singly or in pairs, after 3 d on YM agar at 25℃. The budding was multilateral on a narrow base. After two weeks of culturing on Dalmau plates at 25℃, septate hyphae were formed. In addition, unconjugated asci with spherical ascospores were observed (Fig. 1B-E). This species has been reported to be heterothallic, with one to four hat-shaped ascospores [7].
On the Biolog YT plate, the strain Hyphopichia burtonii KHU210802-01 (GenBank: OR772230) was positive for the oxidation of maltose, maltotriose, sucrose, and α-D-glucose. The strain was found to assimilate several carbon compounds, including maltose, maltotriose, sucrose, and α-D-glucose (Table 2).
Remarks: Pichia burtonii is synonymous to Hyphopichia burtonii; which is a common spoilage yeast that causes food and beverage spoilage, as well as “Chalk mold” defects in partially baked bakery goods, cured meats, and biscuits [8,9]. H. burtonii can cause cutaneous infections in Barbastelle bats [10], although human infections have not yet been reported.
Table 2
Oxidation and assimilation of different carbon sources by yeast species. Analyses were conducted using the YT MicroPlate from Biolog(continued).

Table 2
Oxidation and assimilation of different carbon sources by yeast species. Analyses were conducted using the YT MicroPlate from Biolog(continued).
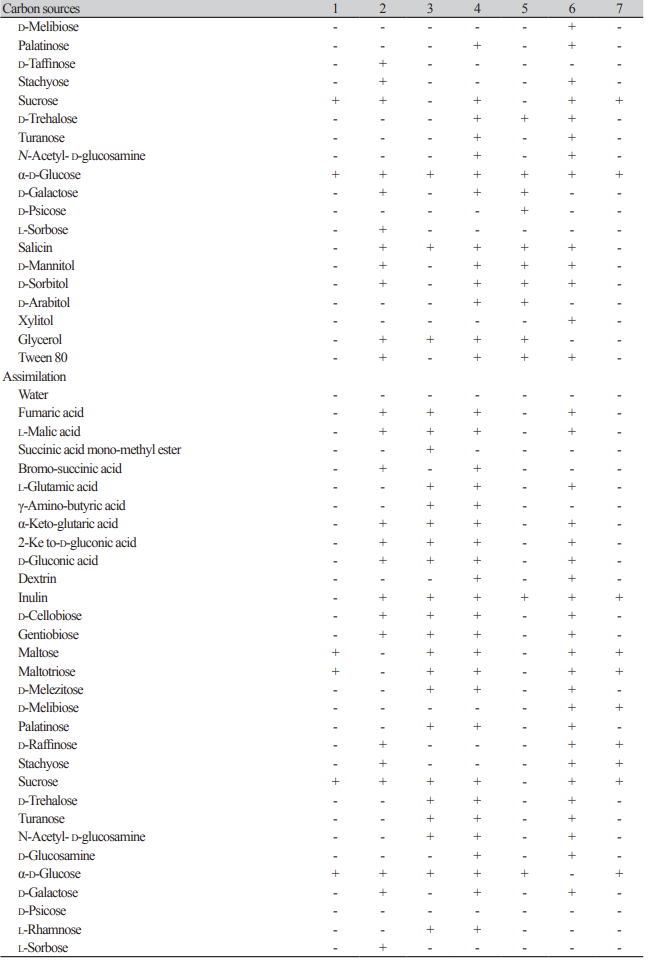
Table 2
Oxidation and assimilation of different carbon sources by yeast species. Analyses were conducted using the YT MicroPlate from Biolog.
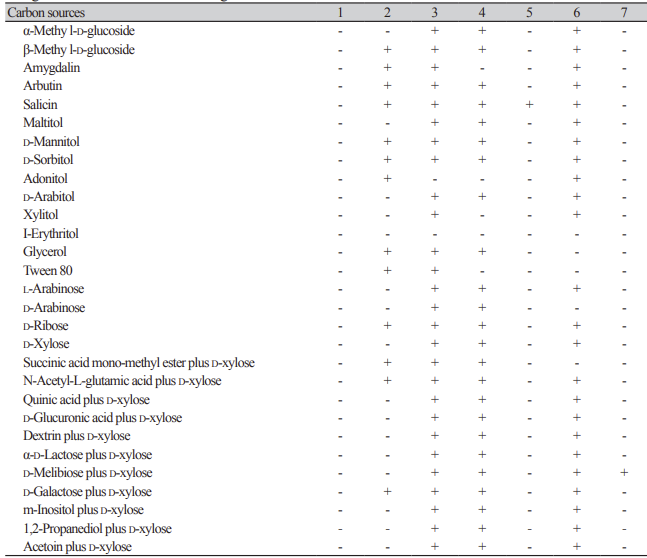
1: Hyphopichia burtonii KHU210802-01; 2: Starmerella sorbosivorans KHU210228-01; 3: Cyberlindnera mycetangii KHU210301-03; 4: Cutaneotrichosporon oleaginosum KHU210228-03; 5: Nakazawaea ernobii KHU210228-04; 6: Pichia kudriazevii KHU210802-02; 7: Schizosaccharomyces japonicus KHU210305-01; +: positive; -: negative.
Starmerella sorbosivorans (S. A. James et al.) C. A. Rosa & Lachance Int. J. Syst. Evol. Microbiol. 68(4): 1333-1343, 2018
Starmerella is a fungal genus that belongs to the order Saccharomycetales. Its relationship with other taxa within the order is uncertain and it has not been definitively placed in any family.
The colonies were smooth, with entire margins, round shapes, and white to ivory in color after 3 d on YM agar at 25℃, while the cells were ovoid to elongated, had a size of 4.1-5.1×5.5-7.4 μm, and occurred either singly or in pairs. The budding was multilateral on a wide base. After two weeks of culturing on Dalmau plates at 25℃, wall-branched pseudohyphae were formed, but hyphae were absent, with roughened and spherical ascospores (Fig. 2B-E). This species has been reported to bud multilaterally [11].
On the Biolog YT plate, the strain Starmerella sorbosivorans KHU210228-01 (GenBank: OR772228) was positive for the oxidation of succinic acid, L-aspartic acid, L-glutamic acid, L-proline, D-gluconic acid, inulin, D-cellobiose, gentiobiose, D-raffinose, stachyose, sucrose, α-D-glucose, D-galactose, L-sorbose, salicin, D-mannitol, D-sorbitol, glycerol, and Tween-80. The strain was also found to assimilate several carbon compounds, including fumaric acid, L-malic acid bromo-succinic acid, α-keto-glutaric acid, 2-keto-Dgluconic acid, D-gluconic acid, inulin, D-cellobiose, gentiobiose, D-raffinose, stachyose, sucrose, α-D-glucose, D-galactose, L-sorbose, β-methyl-D-glucoside, amygdalin, arbutin, salicin, D-mannitol, D-sorbitol, adonitol, glycerol, Tween-80, D-ribose, succinic acid mono-methyl ester plus D-xylose, N-acetyl-L-glutamic acid plus D-xylose, and D-galactose plus D-xylose (Table 2).
Remarks: Several members of the Starmerella clade are associated with flowers and flowervisiting insects, such as bees and bumblebees, which cope well with high-sugar niches. Many strains (species) of the Starmerella clade, including Starmerella bombicola and Candida apicola, are known to produce sophorolipids that are carbohydrate-based, amphiphilic biosurfactants [12,13].
Cyberlindnera mycetangii (Kurtzman) Brysch-Herzb., Dlauchy, M. Seidel & G. Péter, Int. J. Syst. Evol. Microbiol. 71(2): 004477, 2021
Cyberlindnera mycetangii is a fungal genus that belongs to the order Saccharomycetales. The genus Lindnera was originally established by Kurtzman et al. in 2000 to accommodate the species of the clade Pichia americana. Lindnera later became a homonym for a valid plant genus; therefore, the name Cyberlindnera was proposed. Consequently, the recognized species of Lindnera were reassigned to Cyberlindnera in new combinations [14,15].
The colonies had entire margins and round forms and appeared white to ivory in color after 3 d on YM agar at 25℃, while the cells were ovoid to elongated with a size of 2.6-3×7.1-8.9 μm and occurred either singly or in pairs. The bud type exhibited polar budding on a narrow base. Septate hyphae were formed, and globose ascospores and asci were observed (Fig. 3B-E). This species is reported to be a pseudomycelium, in which a true mycelium is formed. Typically, two hat-shaped ascospores were observed [14].
On the Biolog YT plate, the strain KHU210301-03 (GenBank: OR772231) was positive for the oxidation of inulin, α-D-glucose, salicin, and glycerol. It was also found to assimilate several carbon compounds, including fumaric acid, L-malic acid, succinic acid mono-methyl ester, L-glutamic acid, γ-amino-butyric acid, α-ketoglutaric acid, 2-keto-D-gluconic acid, D-gluconic acid, inulin, D-cellobiose, gentiobiose, maltose, maltotriose, -melezitose, palatinose, sucrose, D-trehalose, turanose, N-acetyl-D-glucosamine, α-D-glucose, L-rhamnose, α-methyl-D-glucoside, β-methyl-D-glucoside, amygdalin, arbutin, salicin, maltitol, D-mannitol, D-sorbitol, D-arabitol, xylitol, glycerol, Tween-80, L-arabinose, D-arabinose, D-ribose, D-xylose, succinic acid mono-methyl ester plus D-xylose, N-acetyl-L-glutamic acid plus D-xylose, quinic acid plus D-xylose, D-glucuronic acid plus D-xylose, dextrin plus D-xylose, α-D-lactose plus D-xylose, D-melibiose plus D-xylose, D-galactose plus D-xylose, m-inositol plus D-xylose, 1,2-propanediol plus D-xylose, and acetoin plus D-xylose (Table 2).
Remarks: Although Cyberlindnera mycetangii has been relatively understudied, phylogenetically similar strains, such as C. amicana and C. fabianii, the former acting as a gut microbe of bark beetle (Dendroctonus rhizophagus), are known to degrade various substrates, including starch and lipids, as well as acting as symbiotic fungi for the detoxification of high concentration toxic compounds produced in pine trees [16]; the latter is used in the fermentation of traditional Chinese sweet rice wine to effectively improve the nutritional and physicochemical properties through the solid-state fermentation of barley [17].
Cutaneotrichosporon oleaginosum (J. J. Zhou, S. O. Suh, and Gujjari) Xin Zhan Liu, F. Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81(1): 85147, 2015
Cutaneotrichosporon is a fungal genus belonging to the order Trichosporonaceae; proposed as a monophyletic group consisting of the Cutaneum and Hagleroum clades, as well as Trichosporon guehoae, Cryptococcus cuvatus, Cr. cryanovorans, and Cr. daszewskae [18,19].
The colonies were smooth with entire margins, round forms, and white to ivory in color after 3 d on YM agar at 25℃, while the cells were ovoid to elongated, had a size of 4.9-7.4×8.8-9.5 μm, and occurred either singly or in pairs after 3 d on YM agar at 25℃. The budding was polar on a narrow base. After two weeks of culturing on Dalmau plates at 25℃, pseudohyphae were formed, but hyphae were absent. Globose ascospores and unconjugated asci were also observed (Fig. 4B-E). However, sexual reproduction has not been observed in this species [19]
On the Biolog YT plate, the strain KHU210228-03 (GenBank: OR772232) was positive for the oxidation of succinic acid mono-methyl ester, L-aspartic acid, L-glutamic acid, L-proline, D-gluconic acid, dextrin, inulin, D-cellobiose, gentiobiose, maltose, maltotriose, D-melezitose, palatinose, sucrose, D-trehalose, turanose, N-acetyl-D-glucosamine, α-D-glucose, D-galactose, salicin, D-mannitol, D-sorbitol, D-arabitol, glycerol, and Tween-80. The strain was also found to assimilate several carbon compounds, including fumaric acid, L-malic acid, bromo-succinic acid, L-glutamic acid, γ-amino-butyric acid, α-keto-glutaric acid, 2-keto-D-gluconic acid, D-gluconic acid, dextrin, inulin, D-cellobiose, gentiobiose, maltose, maltotriose, D-melezitose, palatinose, sucrose, D-trehalose, turanose, N-acetyl-D-glucosamine, D-glucosamine, α-D-glucose, D-galactose, L-rhamnose, α-methyl-D-glucoside, β-methyl-D-glucoside, arbutin, salicin, maltitol, D-mannitol, D-sorbitol, D-arabitol, glycerol, L-arabinose, D-arabinose, D-ribose, D-xylose, succinic acid monomethyl ester plus D-xylose, N-acetyl-L-glutamic acid plus D-xylose, quinic acid plus D-xylose, D-glucuronic acid plus D-xylose, dextrin plus D-xylose, α-D-lactose plus D-xylose, D-melibiose plus D-xylose, D-galactose plus D-xylose, m-inositol plus D-xylose, 1,2-propanediol plus D-xylose, and acetoin plus D-xylose (Table 2).
Remarks: Although Cutaneotrichosporon oleaginosum cannot directly degrade chitin, it can utilize GlcNAc, the basic structural unit of chitin, to synthesize lipids [20]. Furthemore, it has also been reported to produce lipids suitable for the production of high-quality biodiesel from waste glycerol, biomass hydrolysates, pumpkin shells, and fruit processing residues [21,22].
Nakazawaea ernobii (J. Lodder & N. J. W. Kreger-van Rij) C. P. Kurtzman & C. J. Robnett, FEMS Yeast Res. 14(7): 1028-1036, 2014
Nakazawaea is a fungal genus that belongs to the order Saccharomycetales. The genus was originally introduced as Torulopsis ernobii by Lodder and Kreger [23], but was later found to be inappropriate and transferred to Candida ernobii by Mey and Yarrow [24]. However, based on sequence differences in the D1/ D2 and ITS regions, it was reclassified as a Nakazawaea species by Kurtzman and Robnett [25].
The colonies had entire margins and round forms and appeared white to ivory in color after 3 d on YM agar at 25℃, while the cells were ovoid to elongated with a size of 3.3-3.5×5.4-6.6 μm and occurred either singly or in pairs. The budding was polar and on a narrow base. After two weeks of culturing on Dalmau plates at 25℃, pseudohyphae were formed, but hyphae were absent. No ascospores or asci were observed (Fig. 5B-E). This species has been reported to be conjugated and ascospore formation has not been observed [25].
On the Biolog YT plate, the strain KHU210228-04 (GenBank: OR772229) was found to be positive for the oxidation of inulin, D-cellobiose, gentiobiose, maltotriose, D-trehalose, α-D-glucose, D-galactose, D-psicose, salicin, D-mannitol, D-sorbitol, D-arabitol, glycerol, and Tween-80. Furthemore, it was also found to assimilate several carbon compounds, including inulin, α-D-glucose, and salicin (Table 2).
Remarks: Nakazawaea ernobii is effective against Diplodia natalensis infection when used in combination with tea polyphenol [26]. It has also shown potential to rapidly degrade diesel oil and can be used for biodiesel purification [27]. Additionally, it can used as a starter for the fermentation of plum brandy [28].
Pichia kudriavzevii Boidin, Pignal & Besson, Bull. trimest. soc. mycol. Fr. 81(4): 589, 1965
Pichia is a genus within the order Saccharomycetales. Pichia kudriavzevii was originally described as Issachenkia orientalis by Kudryavtsev [29]. Although Kergenr-ban Rij subsequently transferred I. orientalis to the species Pichia, Boidein et al. proposed a novel name, Pichia kudriavzevii, due to the fact that Pichia orientalis had already been used as a name. Subsequent analysis of the D1/D2 LSU rRNA gene sequences and multigene analysis confirmed that the Issatchenkia species in fact belonged to the Pichia species [30,31].
The colonies were round with curled margins and appeared white to ivory in color after 3 d on YM agar at 25℃ while the cells were ovoid to elongated. The bud type was a true (septate) hyphae with side branches bearing blastoconidia. After two weeks of culturing on Dalmau plates at 25℃, hyphae were present and globose ascospores and asci were observed (Fig. 6B-E). This species is heterothallic; the ascospores vary from roughened or smooth and spherical to hat-shaped, and the asci may be persistent or deliquescent [5].
On the Biolog YT plate, the strain KHU210802-02 (GenBank: OR772234) was positive for the oxidation of succinic acid, L-aspartic acid, L-glutamic acid, L-proline, D-gluconic acid, dextrin, inulin, D-cellobiose, gentiobiose, maltose, maltotriose, D-melibiose, palatinose, stachyose, sucrose, D-trehalose, turanose, N-acetyl-D-glucosamine, α-D-glucose, salicin, D-mannitol, D-sorbitol, xylitol, and Tween-80. Furthemore, it was also found to assimilate several carbon compounds, including fumaric acid, L-malic acid, L-glutamic acid, α-keto glutaric acid, 2-keto-D-gluconic acid, D-gluconic acid, dextrin, inulin, D-cellobiose, gentiobiose, maltose, maltotriose, D-melezitose, D-melibiose, palatinose, D-raffinose, stachyose, sucrose, D-trehalose, turanose, N-acetyl-D-glucosamine, D-glucosamine, D-galactose, α-methyl-D-glucoside, β-methyl-D-glucoside, amygdalin, arbutin, salicin, maltitol, D-mannitol, D-sorbitol, adonitol, D-arabitol, xylitol, L-arabinose, D-ribose, D-xylose, N-acetyl-L-glutamic acid plus D-xylose, quinic acid plus D-xylose, D-glucuronic acid plus D-xylose, dextrin plus D-xylose, α-D-lactose plus D-xylose, D-melibiose plus D-xylose, D-galactose plus D-xylose, m-inositol plus D-xylose, 1,2-propanediol plus D-xylose, and acetoin plus D-xylose (Table 2).
Remarks: Pichia kudriavzevii and its anamorph Candida krusei are commonly found in soil, fruits, and various natural ferments. Although P. kudriavzevii is not commonly associated with food spoilage, it is frequently isolated from agricultural products and foods. Conversely, C. krusei, is commonly isolated from humans and animals, and is recognized as a pathogen based on clinical evidence provided by Hurley et al. [32]. Pichia kudriavzevii has the potential to be used as a starter for the fermentation of plum brandies [28]. It can positively influence the physical, chemical, and biological properties of soil, leading to improvements in soil quality and crop production [33]. It is also known to exert a good fermentative capacity for the production of gluten-free sourdough when used as a starter [34].
Schizosaccharomyces japonicus Yukawa M, Maki T, Bul. Sci. Fak. Terkultura Kjusu Imp. Univ. 4: 224-242, 1931
Schizosaccharomyces japonicus is a fungal genus that belongs to the order Schizosaccharomycetales. Two varieties exist, namely Sz. japonicus and Sz. versatilis, which were originally distinguished by differences in the ascospore shape and positive iodine reactions in the mature ascospores of Sz. versatilis strains [35,36]. However, high nuclear DNA similarity suggests that varietal designations are unnecessary [37]. Previous studies have reported that a high level of cross-pollination occur between the two varieties [38,39], and that the electrophoretic mobilities of selected enzymes from strains representing both varieties were identical [40].
The colonies had entire margins and irregular forms and appeared ivory in color after 3 d on YM agar at 25℃, while the cells were ovoid to elongated with a size of 5.3-6.8×7.2-10 μm and occurred either singly or in pairs. The budding was multilateral on a wide base. After two weeks of culturing on Dalmau plates at 25℃, hyphae were formed, while ascospores and asci were present at times (Fig. 7B-E). This species has been reported to have asci, which are usually produced by conjugation, as well as ascospores that are globose or short ellipsoidal and seldom cylindrical [5].
On the Biolog YT plate, the strain Schizosaccharomyces japonicus KHU210305-01 (GenBank: OR772227) was positive for the oxidation of inulin, sucrose, and α-D-glucose. Furthemore, it was found to assimilate several carbon compounds, including inulin, maltose, maltotriose, D-melibiose, D-raffinose, stachyose, sucrose, α-D-glucose, and D-melibiose plus D-xylose (Table 2).
Remarks: Schizosaccharomyces japonicus can be used as a fermentation starter in wine and beer production. Their addition during wine fermentation increases the production of beneficial compounds, including acetate esters [41]. During fermentation, polysaccharides are released, which positively contribute to the protein stability of wine [42]. This strain has also been used as a fermentation starter for the traditional Korean rice wine, makgeolli, producing a unique and distinctive flavor profile with abundant ester compounds [43].