Erysiphe powdery mildew on Chloranthus japonicus Siebold (Chloranthaceae) was first described in 1968 as Erysiphe communis f. chloranthi Golovin & Bunkina, based on a specimen collected at Primorsky Krai, in the Russian Far East [1,2]. Braun subsequently raised it to the species level because it is the only powdery mildew known in plants from the family Chloranthaceae, thus being E. chloranthi (Golovin & Bunkina) U. Braun [3]. To date, this species has been recorded on C. japonicus from Japan [4] and Korea [5].
However, the morphological characteristics of E. chloranthi are insufficiently known [2]. Characterization of asexual morphs was neglected in the original description provided by Golovin & Bunkina [1]. Nomura [4] described and illustrated the teleomorph of this species without mentioning its anamorph. Shin [5] described the morphological characteristics of both the anamorph and teleomorph. However, there are no distinctive characteristics that distinguish this species from other related species (formerly E. communis s. lat. or E. polygoni s. lat).
Regarding the molecular data characterizing this species, only one Japanese specimen collected from Chloranthus serratus Roem & Schult has been used to obtain nucleotide sequences of the internal transcribed spacer (ITS) regions, large subunit (LSU), and MCM7 genes of rDNA in phylogenetic studies of Erysiphaceae [6]. Here, we describe the morphological characteristics of the holomorph and indicate the phylogenetic position of the fungus based on newly obtained sequences from Korean specimens.
During our extensive field forays to collect phytopathogenic fungi in Korea, seven samples of C. japonicus infected with powdery mildew were collected and preserved at the Korea University Herbarium (KUS). They were deposited as KUS-F16196 (Jul 6, 1999; Wild Vegetable Experimental Station, Pyeongchang), F18469 (Sep 17, 2001; Experimental Forest of Kangwon National University, Hongcheon), F23725 (Oct 2, 2008; Bukbang-myeon, Hongcheon), F25031 (Jun 29, 2010; Mt. Gongjaksan, Hongcheon), F28725 (Jul 6, 2015; Dongsan-myeon, Chuncheon), F28942 (Oct 15, 2015; Mt. Gariwangsan, Pyeongchang), and F31847 (Jul 7, 2020; Mt. Yumyeongsan, Gapyeong). One sample of C. serratus infected with powdery mildew was collected and preserved as KUS-F13093 (Sep 28, 1994, Seongsanmyeon, Gangneung) but was not included in the present study because of the poor condition of the sample.
Detailed morphological characteristics of the powdery mildew were observed using an Olympus BX50 microscope (Olympus, Tokyo, Japan). Photomicrographs were captured using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). All microscopic measurements and characterizations were performed using dried herbarium material. Photomicrographs of anamorphic structures were obtained using the lactic acid technique outlined by Shin & La [7], and an NaOH solution (3% w/v) was used to examine sexual morphs. Thirty measurements were performed for each diagnostic structure.
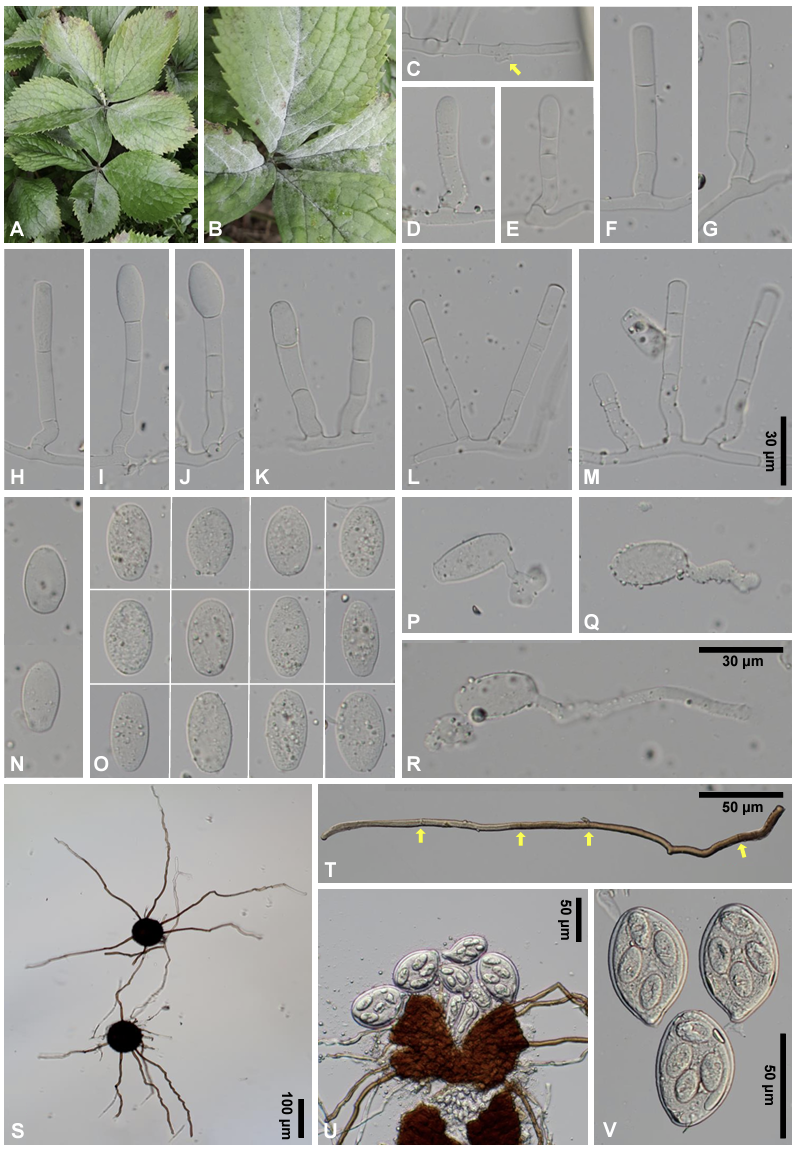
Mycelial mats with conidiophores and conidia were mostly epigenous, less frequently amphigenous, and occasionally cauligenous, forming conspicuous white colonies on the upper leaf surface and thinly effused on the lower leaf surface (Figs. 1A and B). In the later stages of the disease, chasmothecia formed on both sides of the leaves and occasionally on the stems. Appressoria on the mycelium were moderately lobed to multilobed, solitary or in opposite pairs, and 3-8 μm wide (Fig. 1C). Conidiophores were 38-116×7-9 μm, and produced conidia singly, followed by (1-)2-3 cells (Figs. 1D-M). Foot-cells of the conidiophores were straight or f lexuous, cylindrical, and 12-33 μm long. Conidia were hyaline, cylindric oval to ellipsoid, variable in shape and size, measured 24-42×14-18 μm (length/width ratio=1.6-2.8), and lacked conspicuous fibrosin bodies (Fig. 1O). Primary conidia were generally smaller than the conidia and were characterized by a round apex and subtruncate base (Fig. 1N). Germ tubes were produced on the perihilar position of the conidia and were short, with a lobed apex or long hyphae (Figs. 1P-R). Chasmothecia were amphigenous, cauligenous, scattered to gregarious, dark brown, spherical, and 76-94 μm in diameter (Fig. 1S). Chasmothecial appendages were positioned on the lower half of the chasmothecium, numerous, 3-16 in number, mostly fewer than 10, mycelioid, usually simple, rarely irregularly branched, interlaced with the aerial hyphae and each other, 0.5-6 times as long as the chasmothecial diameter, 4-7 µm wide, dark brown at the base and paler upwards, and 1-7-septate (Figs. 1S-U). Asci were 4-6 in a chasmothecium, obovoid, saccate, shortstalked, and 48-64×32-40 µm (Fig. 1V). Ascospores were (3-)4-6 in an ascus, oval, and 20-26×12-15 µm (Fig. 1V).
Fig. 1
Erysiphe chloranthi, a powdery mildew found on Chloranthus japonicus. A: Powdery mildew infections on the leaves and inflorescence. B: Close-up view of affected leaves. C: Appressoria (arrow) on the hypha. D and E: Primary conidiophores characterized by the round apex of the uppermost cell. F-M: Diversity of conidiophores with straight or flexuous foot-cells. The scale bar in M has been applied to C-M. N: Primary conidia characterized by the round apex and subtruncate base, thus an asymmetric shape. O: Diversity of conidia exhibiting various shapes and a wide range of length/width ratios. P-R: Conidia in germination. The scale bar in R has been applied to N-R. S: Chasmothecia with appendages. T: Chasmothecial appendage. Arrows indicate the positions of septa. U: A crushed chasmothecium exhibiting six asci. V: Asci containing four ascospores each. The asci and ascospores were slightly pressed to show the shape clearly.
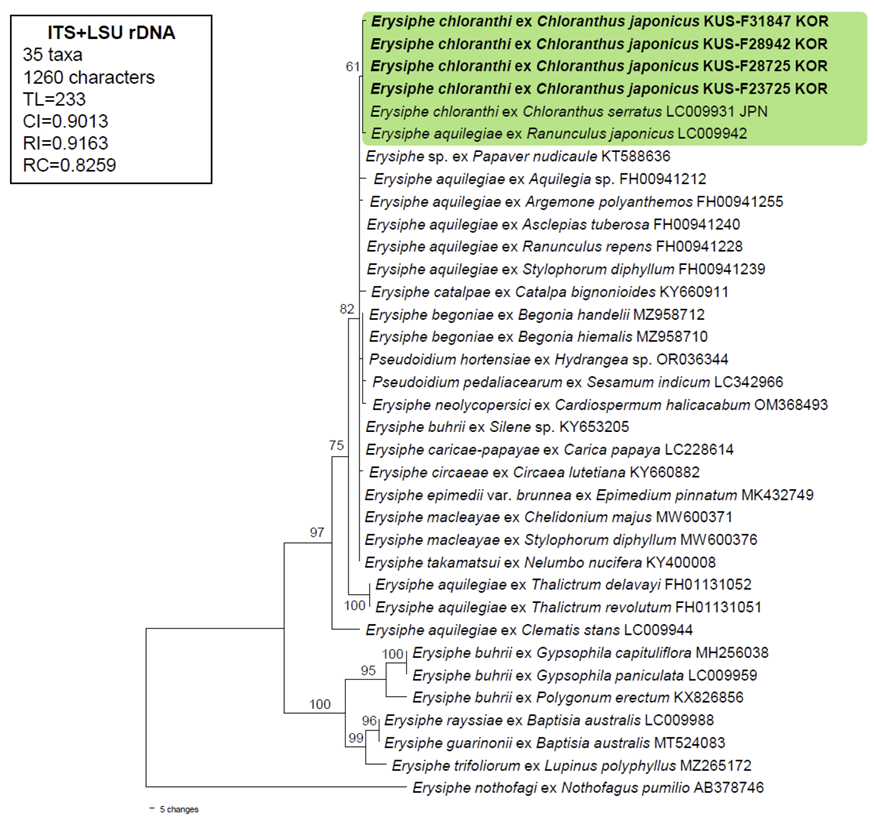
DNA extraction, PCR, and sequencing were performed as previously described by Choi et al. [8]. The nucleotide sequences of the ITS regions, including the 5.8S gene and LSU gene of rDNA, were determined from KUS-F31847, F28942, F28725, and F23725. The resulting forward and reverse sequences were assembled in MEGA11 and deposited in GenBank under the following accession numbers: OR512525, OR512526, OR512527, and OR512528 for ITS region and OR512529, OR512530, OR512532, and OR512533 for LSU. To perform phylogenetic analysis, a combined dataset of ITS+LSU using 35 sequences of the genus Erysiphe was created and aligned using the MUSCLE command in MEGA11 [9]. Erysiphe nothofagi (Thaxt.) U. Braun & S. Takam. were selected as outgroups. A phylogenetic tree was constructed in PAUP* 4.0a based on the maximum parsimony method, with the application of the heuristic search option using the ʻtree-bisection-reconstruction’ (TBR) algorithm from 100 random sequence additions to find the global optimum tree. All sites were treated as unordered and unweighted, and gaps were treated as missing data [10]. The strength of the internal branches in the resulting tree was tested using 1,000 replications in bootstrap (BS) analysis. Tree scores, such as tree length, consistency index (CI), retention index (RI), and rescaled consistency index (RC) were calculated. The data matrix consisted of 35 sequences and 1,260 characters, of which 109 (8.65%) were variable and parsimony-uninformative, and 70 (5.55%) were informative for parsimony analysis.
With regard to morphological characteristics, E. chloranthi of the Korean specimens is characterized by very short foot-cells in the conidiophores. Braun & Cook [2] provided the data of foot-cells as (15-)20-40 ×7-11 µm, which was obtained from many reports on E. aquilegiae s. lat. Compared to these data, 12-33 ×7-9 μm for foot-cells in the Korean specimens appears to be a key characteristic for this species. The other characteristics of the conidiophores and conidia were in the scope of E. aquilegiae s. lat. [2]. The morphological characteristics noted for the chasmothecia, chasmothecial appendages, asci, and ascospores were also within the scope of E. aquilegiae.
Comparison of newly obtained sequences with reference sequences in GenBank using BLASTn search showed 99.82% identity with Erysiphe sp. (KT588636), E. aquilegiae (MN108157, KY660916, KY653201-KY653204), E. catalpae (KY660911), and E. buhri (KY653205). The four specimens collected from different locations in Korea shared 100% identity in nucleotide sequences of ITS region and LSU. In the resulting phylogenetic tree, these four sequences were clustered with sequences of E. chloranthi (1,344 bp) and E. aquilegiae (1,344 bp) on Ranunculus in a small cluster within the aquilegiae clade and shared 100% identity in the ITS region and LSU sequences (Fig. 2), suggesting Erysiphe powdery mildew on Chloranthus is derived from the same ancestor with E. aquilegiae. The bootstrap value of this branch was 61%, which is lower than 70. Our results are in good agreement with those of the phylogenetic analysis of Erysiphe sect. Microsphaera conducted by Takamatsu et al. [11], where E. chloranthi was placed in the aquilegiae clade with E. catalpae, E. euphorbiae, E. macleayae, E. knautiae, E. coriariae, E. circaeae, E. sedi, E. hommae, E. pileae, E. takamatsui, Pseudoidium neolycopersici, Ps. hortensiae, and Ps. boroniae.
Fig. 2
A maximum parsimony tree of Erysiphe chloranthi, which is placed within the E. aquilegiae complex, was derived from a combined data matrix of the ITS+LSU sequences based on Takamatsu et al. (2015). The isolates obtained in this study are shown in bold. Bootstrap values (>60%) are indicated on the related branches. ITS, internal transcribed spacer; LSU, large subunit; TL, tree length; CI, consistency index; RI, retention index; RC, rescaled consistency index.
Chloranthaceae is the only family in the order Chloranthales and is considered one of the early diverging angiosperm lineages [12]. Recent molecular phylogenetic analyses have revealed that the family is not closely related to any other families of flowering plants [13]. Currently, four genera, Ascarina, Chloranthus, Hedyosmum, and Sarcandra, comprise this family. Of these, Chloranthus is the only taxon known to be infected with powdery mildew [2,14]. Considering host-parasite co-speciation in the Erysiphaceae (powdery mildews) [2,4,5,11], E. chloranthi can be maintained as a distinct species until the complexity or ambiguity of the E. aquilegiae complex is solved.
Based on the morphological circumscriptions and the results of molecular phylogenetic analyses, the powdery mildew on C. japonicus was confirmed to be E. chloranthi. This is a confirmed report of E. chloranthi on C. japonicus, providing the holomorph morphology and the first sequence data for this powdery mildew from Korea. This study may help elucidate the identity of this species and clarify the ambiguity of the E. aquilegiae complex.


