INTRODUCTION
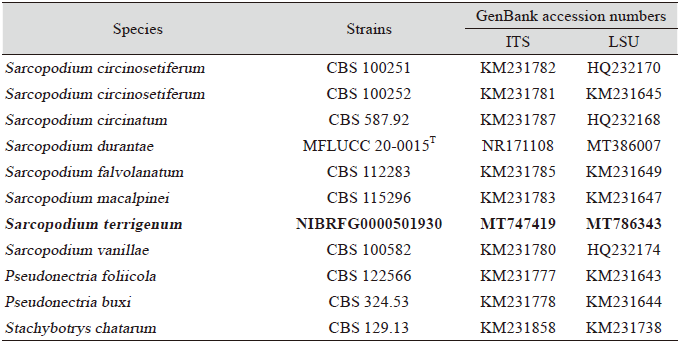
The genus Sarcopodium belonging to the family Nectriaceae, order Hypocreales, class Sordariomycetes, subphylum Pezizomycotina, and phylum Ascomycota, was first described in 1818, with Sarcopodium circinatum as the type species. Since then, the genus has had numerous names (obsolete synonyms: Tricholeconium, Periolopsis, Actinostilbe, Kutilakesa, and Kutilakesopsis) [1-3]. Currently, the genus has 30 members according to the MycoBank database (http://www.mycobank.org). Sarcopodium is characterized by the formation of sporodochia with curled, sterile, ornamented, dark setae arising from branched, septate conidiophores, and hyaline phialoconidia. The setae are simple, septate, rarely branched, smooth or verruculose, and straight or circinate; they can be rounded in exceptional cases. The conidia are aggregated in slimy masses, hyaline, straight, and cylindrical to ellipsoid [1,3,4]. Species of this genus are found on wood, dead herbaceous stems, human skin, and plant leaves [4-6]. Recently, Sarcopodium sp. was discovered in the deep sea [7].
During the screening of the novel and unreported fungi in the Republic of Korea, the fungal strain YW23-8 was isolated from a soil sample. Based on cultural, morphological, and molecular characteristics, the isolate represents an undescribed species of the genus Sarcopodium. In this study, the isolated fungus is described and illustrated as a novel species.
MATERIALS AND METHODS
Soil sampling and fungal isolation
Strain YW23-8 was isolated from a soil sample collected in Korea (37°16′30.7″N, 128°31′29.9″E). The sample was serially diluted in distilled water, then 100 μL of each dilution was spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates. The fungal colonies were selected and isolated separately based on cultural characteristics after incubation for 2-3 days at 25℃, by sub-culturing under the same conditions.
Morphological characterization
To study the morphology, YW23-8 was cultured on three different media, PDA, malt extract agar (MEA; Difco, Detroit, MI, USA), and oatmeal agar (OA; Difco, Detroit, MI, USA), for 5 days at 25℃ [1]. Following incubation, the colony characteristics including color, shape, and size were recorded. A light microscope was utilized to observe the morphological structure of the strain (BX-50; Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Genomic DNA of strain YW23-8 was extracted using a DNA extraction kit (BIOFACT, Daejeon, Korea) according to the manufacturer’s instructions. Molecular identification based on PCR amplification was conducted using the internal transcribed spacer (ITS) regions and partial sequences of the large subunit of the nuclear ribosomal RNA (LSU) gene to determine the genus and species. The primers ITS1F and ITS4 [8,9] were used to amplify the ITS regions, and the LSU gene was amplified using the primer pair V9G/LR5 [10,11]. The PCR conditions for ITS included an initial elongation step for 2 min at 94℃; followed by 35 cycles of 30 s at 94℃, 45 s at 55℃, 90 s at 72℃, and a final elongation step for 5 min at 72℃. The conditions for LSU were 3 min at 94℃; followed by 35 cycles of 35 s at 94℃, 1 min at 49℃, 2 min at 72℃, and finally 5 min at 72℃. The PCR products were purified using EXO-SAP (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced (Solgent, Daejeon, Korea).
Phylogenetic analysis
The generated DNA sequences were compiled and assembled in BioEdit [12] and compared with closely related species retrieved from National Center for Biotechnology Information (NCBI). The ITS and LSU sequences of related taxa were obtained from GenBank (Table 1). These sequences were combined and aligned using Clustal X [13]. A phylogenetic tree based on concatenated ITS and LSU sequences was constructed with MEGA 7 program [14] using the neighbor-joining (NJ) method. A bootstrap analysis with 1,000 replicates was used to calculate the confidence levels of branches [15].
RESULTS AND DISCUSSION
Taxonomy
Sarcopodium terrigenum S.E. Nabil, S.Y. Lee & H.Y. Jung sp. nov. (Fig. 1)
MycoBank: MB839077
Etymology: This refers to the soil, the substrate from which the type strain was isolated.
Typus: The culture was isolated in Korea (37°16′30.7″N, 128°31′29.9″E). The stock culture (NIBRFG0000501930) was deposited at the National Institute of Biological Resources as a metabolically inactive culture.
Cultural and morphological characteristics
YW23-8 was cultured on PDA, OA, and MEA at 25℃ to observe its morphological structure (Fig. 1A-C). The colony on PDA, reaching 65 mm in 15 days, was circular, slightly raised, and curled with a rough surface. The color of the colony was initially yellowish-white, and over time it became darker and brownish-yellow from the center outward, with yellow on the reverse side. The milky white droplets were globular and smooth, and they arose from the ends of the conidiophores (Fig. 1D). The setae were always straight, thin-walled, and septate, with a swollen setal apex forming a light pulp shape end, generally 2.5–3.5 µm and up to 6 µm wide at the apex (Fig. 1F). The conidia aggregated in slimy masses and were spherical to oval, hyaline, smooth, thin-walled, and aseptate, with dimensions of 4.7–7.0×4.5–5.0 µm, similar to those of the closest related species S. circinosetiferum (Fig. 1H).
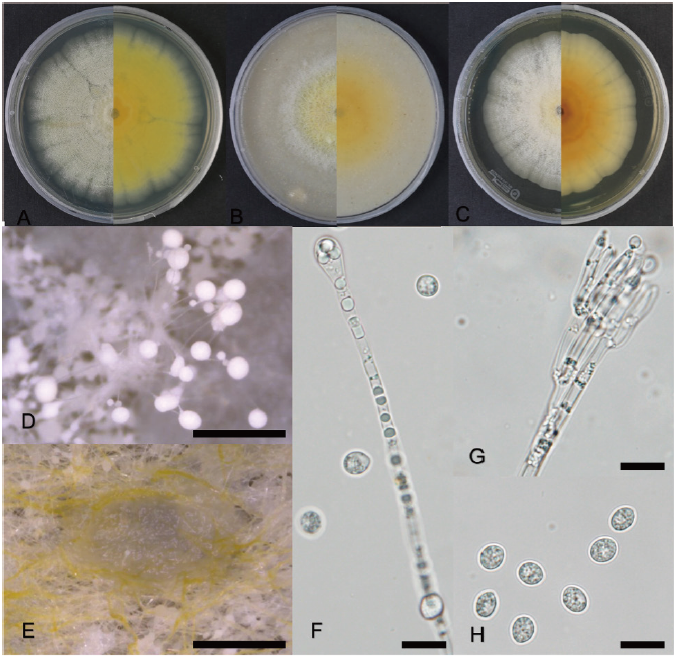
Fig. 1. Cultural and morphological characteristics of Sarcopodium terrigenum YW23- 8. A: Colony on potato dextrose agar. B: Colony on oatmeal agar. C: Colony on malt extract agar. D: Milky watery droplets on potato dextrose agar plate. E: Mycelia on potato dextrose agar. F: Setae. G: Conidiophores. H: Conidia. Scale bars: D, E=500 μm; F-H=10 μm.
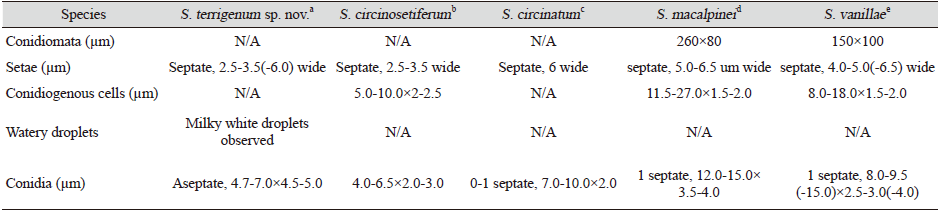
Note: The species S. macalpinei (260×80 µm) and S. vanilla (150×100 µm) produce conidiomata, which are absent in S. circinosetiferum, S. circinatum, and the novel isolate YW23-8. The species S. circinosetiferum (septate), S. circinatum (0–1 septate), S. macalpinei (1 septate), and S. vanillae (1 septate) produce septate conidia, whereas YW23-8 produces aseptate conidia (Table 2). Sarcopodium circinosetiferum, S. circinatum, S. macalpinei and S. vanillae have different types of conidial structures, but YW23-8 produced spherical to oval conidia (Table 2). Moreover, in contrast to S. circinosetiferum, S. circinatum, S. macalpinei and S. vanilla, YW23-8 produced numerous milky white watery droplets. A comparison with the nearest species of the genus showed that YW23-8 is different due to its conidial structures, the absence of septation in the conidia, and the presence of numerous milky white watery droplets. Thus, the morphology of YW23-8 is significantly different from that of its closest species, the previously described Sarcopodium species.
Phylogenetic analysis
The sequences of the ITS regions of YW23-8 showed a similarity of 99.5-100% to those of S. circinosetiferum PSN02 (KT184722) and S. circinosetiferum PIP09 (KT184703). However, YW23-8 shared only 93.5% identity with S. circinosetiferum CBS 100252T (KM231781), which is the type species. Identification of the closest taxonomic relatives using the LSU gene sequence revealed a high similarity of 98–99% between the isolate and S. macalpinei (CBS 128372, CBS 128371), S. flavolanatum (CBS 230.31, CBS 172.25), and S. vanillae (MFLUCC 17-2597). These results clearly indicate that a comparative analysis based on the sequence of one LSU gene or ITS region would not precisely identify the novel fungal strain. Therefore, phylogenetic analysis using concatenated ITS and LSU gene sequences were performed for accurate differentiation. The NJ phylogenetic tree based on the concatenated ITS and LSU gene sequences were clearly demonstrated that the strain S. terrigenum YW23-8 occupied a distinct position among the closest Sarcopodium species (Fig. 2). Accordingly, YW23-8 represents a single, novel, phylogenetically distinct species. Sarcopodium species have been reported in various countries, habitats, and substrates, including S. circinatum (soil, Costa Rica; leaf litter, Brazil), S. circinosetiferum (soil, Argentina), S. flavolanatum (evergreen tree, Theobroma gileri, Ecuador; decaying wood, China), S. macalpinei (evergreen shrub, Viburnum odoratissimum, Hong Kong), and S. vanillae (Anthurium sp., Ecuador) [1]. The most recognized species in the genus, S. araliae, S. circinatum (the genus type species), S. oculorum and S. tortuosum, are relatively common plant pathogens. Moreover, S. oculorum, named after the infection site, was reported to cause corneal ulcers in humans [5]. Thus, according to the phylogenetic analysis and morphological observation, strain YW23-8 is distinct from the previously identified Sarcopodium species. It should be placed in the genus as a novel species for which we propose the name Sarcopodium terrigenum sp. nov. Since comprehensive information is still lacking, further investigation is required to discover the habitats, geographic distribution, and pathogenic potential of this new fungal species.
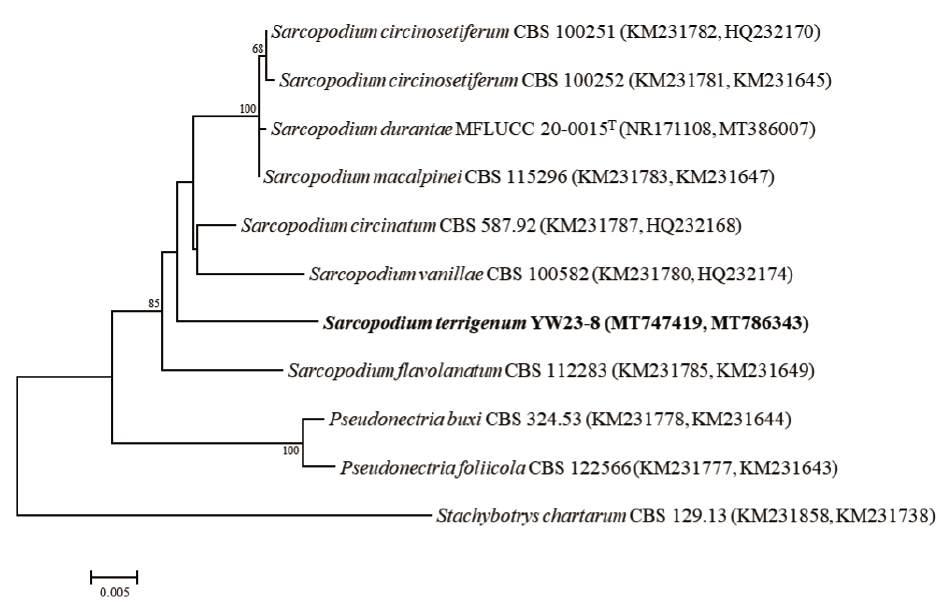
Fig. 2. Neighbor-joining phylogenetic tree based on the concatenated internal transcribed spacer regions and the partial sequence of the large subunit (LSU) rRNA gene, showing the phylogenetic position of the novel strain YW23-8 among the closest Sarcopodium species. The numbers above the branches represent the bootstrap values obtained for 1,000 replicates (values <60% not shown). The strain isolated in this study is indicated in bold. Scale bar: 0.005 substitutions per nucleotide position.