Introduction
Truffles are a group of fungi that produce hypogeous ascocarps; they belong to the genus Tuber (Ascomycota, Pezizales) [1]. Tuber spp. have ectomycorrhizal (ECM) associations with woody plants such as Quercus, Pinus, and Corylus [2,3]. Truffles have been widely used as edible mushrooms in Europe for a long time owing to their unique fragrance and flavor [1].
Tuber himalayense B.C. Zhang & Minter was first reported in 1988 [4] and its morphological characteristics were largely consistent with those of T. indicum, known as Asian black truffle; furthermore, it shared morphological and phylogenetical similarities with European black truffle Tuber melanosporum. However, the fruiting bodies of T. himalayense are smaller than those of T. indicum and polygonal warts attached to the outer side of the peridium has been recorded to be regular compared to those of T. indicum [3,4]. As T. himalayense and T. indicum are systematically very close, their taxonomic distinction has not been clarified and the two species have been considered as one or as T. indicum complex [5]. Recently, T. himalayense and T. indicum have been distinguished at the species level using DNA sequences of the MAT genes, which determine the mating type of Tuber spp. [6,7].
The ascoma of only two Tuber spp. (Tuber aestivum subsp. uncinatum and Tuber huidongense) have been reported in Korea [8, 9]. Herein, we describe the morphological characteristics of the fruiting bodies of T. himalayense found in the rhizosphere of Quercus dentata Thunb. (an oak tree) in Korea and their mycorrhizal roots. We determined whether T. himalayense systematically differs from T. indicum using several molecular markers, including the MAT genes.
Materials and Methods
The ascomata of T. himalayense were collected from the rhizosphere of Q. dentata in Danyang, Korea. The morphological characteristics of ascoma, asci, and ascospores were examined under a light microscope. For molecular identification, genomic DNA was extracted from the ascoma and ITS1/ITS4 primers [10] and LR0R/LR16 primers were used to amplify internal transcribed spacer (ITS) regions and large subunits (LSU) of ribosomal DNA (rDNA) [11], respectively. EF1α Tuber-f and EF1α Tuber-r, and Tuber-r [12] were also used to amplify the translation elongation factor 1-α. Additionally, the i3/il2 primers and i5/il3 primers [7] were used to amplify MAT 1-1-1 and MAT 1-2-1 DNA for phylogenetic analysis with T. indicum. After PCR amplification, DNA sequences were analysed (SolGent Co., Ltd., Daejeon, Korea). Sequence similarities were determined using BLAST available from the National Center for Biological Information (NCBI). Phylogenetic trees were constructed using the neighbor-joining method with MEGA7 software [13].
The roots of Q. dentata were collected from the same sites in Danyang where the ascoma were collected. The morphological and anatomical characteristic of ECM were examined under a stereomicroscope and light microscopes. Genomic DNA was extracted from the ECM root tips using DiaStarTM Direct Lysis Buffer (SolGent Co., Ltd., Daejeon, Korea). The ITS region of the rDNA was amplified using the ITS1F/ITS4 primers and identified through phylogenetic analysis of the sequences.
Results and Discussion
Taxonomy
Tuber himalayense B.C. Zhang & Minter, Transactions of the British Mycological Society 91 (4): 595 (1988) [MB#134661] (Fig. 1; Table 1)
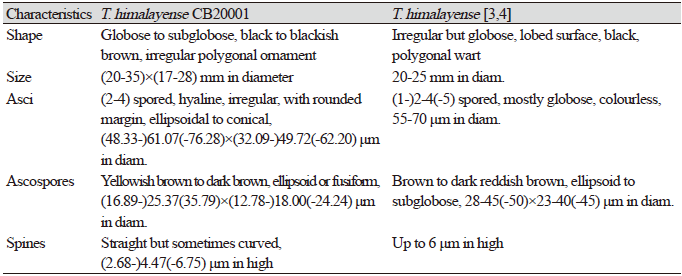
Globose to subglobose, black to blackish brown, 20–35×17–28 mm in diameter (Figs. 1A and 1B), polygonal ornamentation on the surface (Fig. 1C), 0.3-0.5×0.2-0.3 mm in diameter. Peridium divided into two distinct layers: an outer layer, dark brown to black, and (222.90-)267.51(-297.29) μm in thickness, and an inner layer, purplish brown to reddish brown, and (113.71-)135.01(-165.16) μm in thickness. Gleba initially greyish beige and dark brown in the part with mature ascospores (Fig. 1D). Asci hyaline, irregular, with rounded margin, ellipsoidal to conical, differing with the number of ascospores (2-4) in each ascus, (48.33-) 61.07 (-76.28) × (32.09-) 49.72 (-62.20) μm in diam. (Figs. 1E and 1F). Ascospore yellowish brown to dark brown, ellipsoid or fusiform, and (16.89-) 25.37 (-35.79) × (12.78-) 18.00 (-24.24) μm in diameter (Figs. 1G and 1H). The reticular structure on the surface of the mature ascospore resembles a turtle shell, with several spines on the external surface. These spines narrowed away from the basal structure and were usually straight although a few were curved, and (2.68-) 4.47 (-6.75) μm in length.
Specimen examined. Maepo-eup, Danyang-gun, Chungcheongbuk-do, Korea, October 16, 2020, Tuber himalayense B.C. Zhang & Minter, isolated from the rhizosphere of Quercus dentata, strain CB20001. GenBank accession numbers, MW393547 (ITS rDNA from ascoma), MW386847 (LSU rDNA from ascoma), MW810351(TEF1 from ascoma) and MW403878 (ITS rDNA from ECM roots).
Phylogenetic analysis
The phylogenetic analysis showed that the ascoma of CB20001 shared similarity of 98.87% with T. himalayense AB553388.1 for the ITS regions, 98.68% with T. himalayense AB553517.1 for the LSU regions, and 99.88% with T. himalayense AB553537.1 for the TEF1 regions. Phylogenetic analysis showed that the sequences for CB20001 was included in the same clade as T. himalayense (Fig. 2). Additionally, the sequence analysis of the MAT genes showed 100% similarity with T. himalayense LC312350.1 for the MAT 1-1-1 region and 100% similarity with T. himalayense LC312317.1 for the MAT 1-2-1 region. The phylogenetic analysis confirmed that these sequences formed a distinct clade with the sequences of T. indicum (Fig. 3).
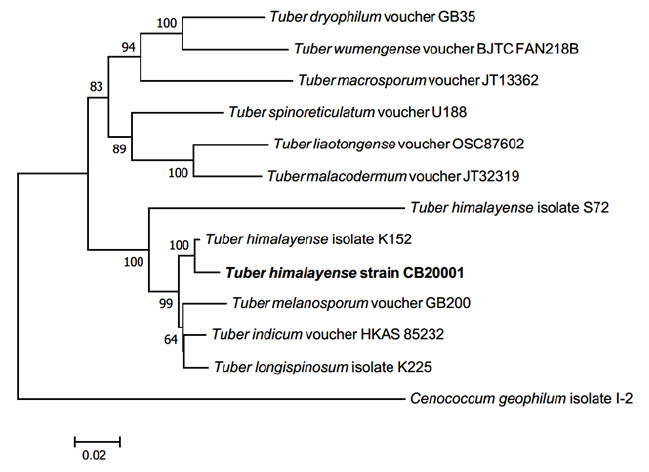
Fig. 2. Neighbor-joining phylogenetic tree of T. himalyense CB20001 ascoma based on concatenated alignment of internal transcribed spacer (ITS), large subunit (LSU) rDNA, and translation elongation factor 1-α (TEF1) DNA sequences. Cenococcum geophilum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicated). The sequence from the present study is shown in bold.
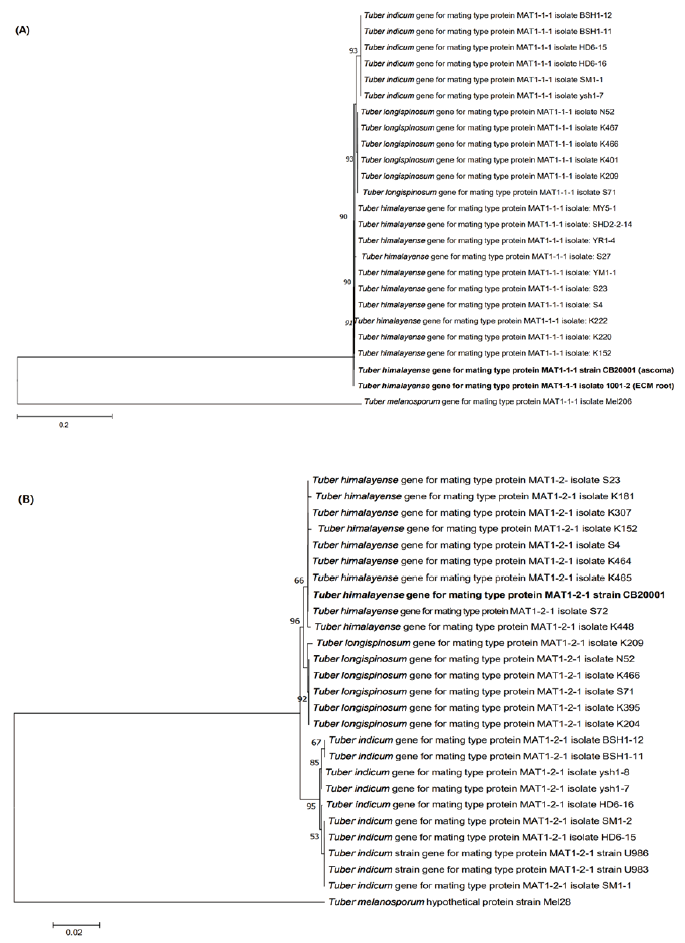
Fig. 3. Neighbor-joining phylogenetic tree of T. himalayense based on the alignment of MAT 1-1-1 (A) and MAT 1-2-1 (B) sequences. Tuber melanosporum was used as the outgroup in both (A) and (B). Numbers on branches indicate bootstrap values (1,000 replicate). The sequences from the present study are shown in bold.
Ectomycorrhiza of T. himalayense x Q. dentata
The ECM root tips were straight, velvety and brown to reddish brown (rarely red) (Figs. 4A-C). The cross section of the ECM showed the fungal mantle and Hartig net (Figs. 4D and 4E). The fungal mantle layer had an interlocking irregular synenchyma (Fig. 4F). The BLAST results showed that the ITS sequence of the ECM root tip is closely related to that of T. himalayense AB553393.1 (99.66% similarity); thus, it was identified as T. himalayense (Fig. 5).
The phylogenetic position of T. himalayense has not been clear due to its highly similar morphological characteristic with T. indicum. Chen et al. [14] classified T. himalayense and T. indicum into a T. indicum complex based on the phylogenetic analysis of LSU rDNA and beta-tubulin (TUB) DNA, whereas Qiao et al. [15] suggested classifying T. indicum and T. himalayense into different groups based on the analysis using simple sequence marker (SSRs). Recently, phylogenetic analysis using ITS, TUB, TEF1, and mating genes, MAT1-1-1 and MAT1-2-1, showed that the two species can be divided into separate clades and a new species, T. longispinosum was positioned between the clades Kinoshita et al. [9]. The ascoma of CB20001 collected from the rhizosphere of the oak tree was identified in this study using the ITS, LSU, and TEF1 regions and MAT genes, and the phylogenetic analysis validated the identification of CB20001 as T. himalayense. Additionally, the morphological characteristics of mycorrhizas of T. himalayense associated with Q. dentata were confirmed. The host plants of T. himalayense include Quercus acutissima Carruth., Quercus serrata Thunb. ex. Murray, and Pinus densiflora Siebold & Zucc. [9], however, in this study, Q. dentata was confirmed as a host plant of T. himalayense.
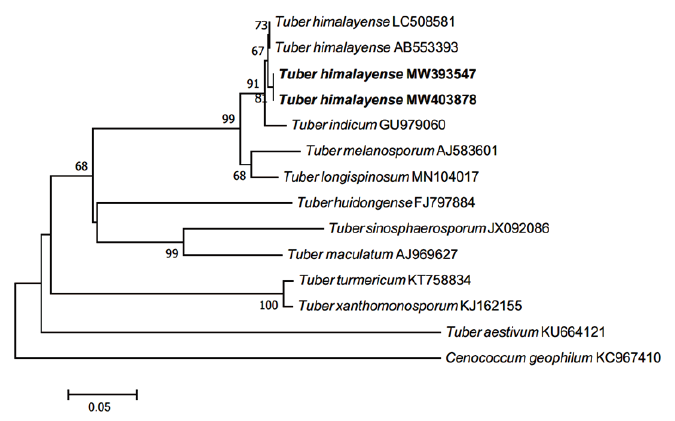
Fig. 5. Neighbor-joining phylogenetic tree of T. himalayense based on the alignment of internal transcribed spacer (ITS) rDNA sequences obtained from ectomycorrhizal root tips (MW403878) and ascoma of CB20001 (MW393547) in this study. Cenococcum geophilum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Sequences from the present study are shown in bold.